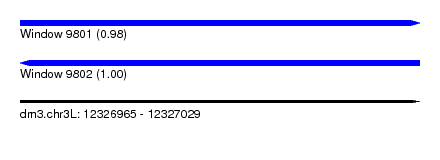
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,326,965 – 12,327,029 |
| Length | 64 |
| Max. P | 0.997682 |

| Location | 12,326,965 – 12,327,029 |
|---|---|
| Length | 64 |
| Sequences | 10 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Shannon entropy | 0.37002 |
| G+C content | 0.51914 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -12.61 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
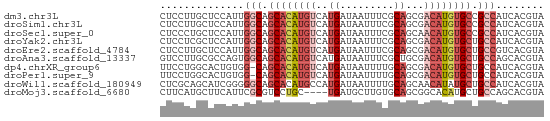
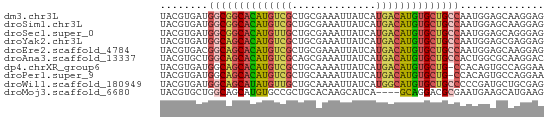
>dm3.chr3L 12326965 64 + 24543557 CUCCUUGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCGACAUGUGCCGCCAUCACGUA .............((((.(((((((((.((.........))..))))))))).))))....... ( -21.90, z-score = -2.99, R) >droSim1.chr3L 11705285 64 + 22553184 CUCCUUGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCGACAUGUGCCGCCAUCACGUA .............((((.(((((((((.((.........))..))))))))).))))....... ( -21.90, z-score = -2.99, R) >droSec1.super_0 4511823 64 + 21120651 CUCCCUGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCAACAUGUGCCGCCAUCACGUA .............((((.((((((((..((.........))...)))))))).))))....... ( -18.30, z-score = -2.05, R) >droYak2.chr3L 12381725 64 + 24197627 CUCCUCGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCGACAUGUGCUGCCAUCACGUA .............((((((((((((((.((.........))..))))))))))))))....... ( -26.50, z-score = -4.66, R) >droEre2.scaffold_4784 12322775 64 + 25762168 CUCCUUGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCGACAUGUGCUGCCGUCACGUA ..............(((((((((((((.((.........))..)))))))))))))........ ( -25.80, z-score = -3.97, R) >droAna3.scaffold_13337 21971410 64 - 23293914 GUCCUUGCGCCAGUGGCAGCACAUGUCAUGAUAAUUUCGCUGCGACAUGUGCUGCCAGCACGUA .....((((....((((((((((((((.(((.....)))....))))))))))))))...)))) ( -28.10, z-score = -3.02, R) >dp4.chrXR_group6 2683588 63 + 13314419 UUCCUGGCACUGUGG-CAGCACAUGUCAUGAUAAUUUUGCAGCGACAUGUGCUGCCAUCACGUA ......((...((((-(((((((((((.((.........))..)))))))))))))))...)). ( -28.00, z-score = -3.78, R) >droPer1.super_9 979720 63 + 3637205 UUCCUGGCACUGUGG-CAGCACAUGUCAUGAUAAUUUUGCAGCGACAUGUGCUGCCAUCACGUA ......((...((((-(((((((((((.((.........))..)))))))))))))))...)). ( -28.00, z-score = -3.78, R) >droWil1.scaffold_180949 2265781 64 - 6375548 CUCGCAGCAUCGGGGGCAGCACAUGCCAUGAUAAUUUUGCAGCAACAUAUGCUGCCAUCACGUA ......((((((..((((.....)))).))))......((((((.....))))))......)). ( -17.50, z-score = -0.24, R) >droMoj3.scaffold_6680 5935054 60 + 24764193 CUUCAUGCUUCAUUCGCGUCCUGC----UGAUGCUUGUGCAGCGGCACAUGCUGCCAGCACGUA .....(((.......(((((....----.)))))..((((.(((((....)))))..))))))) ( -19.30, z-score = -0.34, R) >consensus CUCCUUGCUCCAUUGGCAGCACAUGUCAUGAUAAUUUCGCAGCGACAUGUGCUGCCAUCACGUA ..............(((.((((((((..((.........))...)))))))).)))........ (-12.61 = -13.37 + 0.76)
| Location | 12,326,965 – 12,327,029 |
|---|---|
| Length | 64 |
| Sequences | 10 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Shannon entropy | 0.37002 |
| G+C content | 0.51914 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -19.75 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
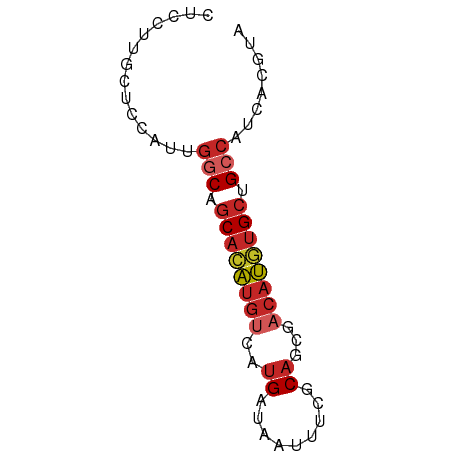
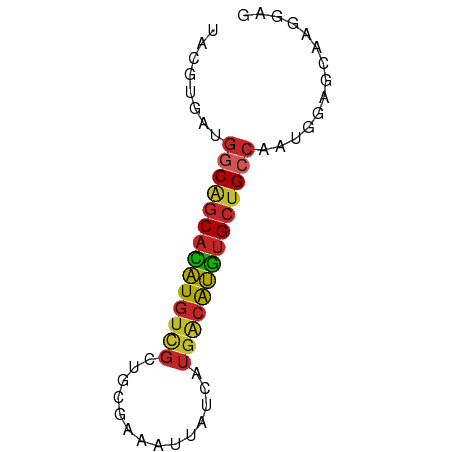
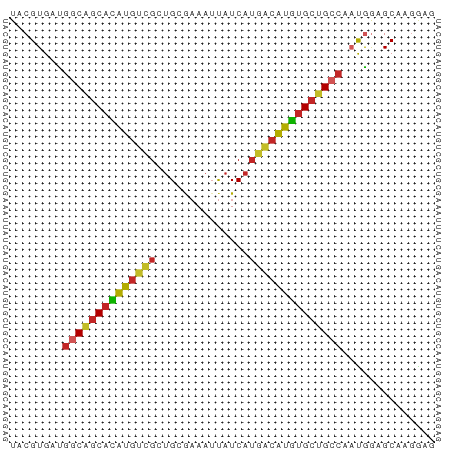
>dm3.chr3L 12326965 64 - 24543557 UACGUGAUGGCGGCACAUGUCGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCAAGGAG ..(((..(((((((((((((((.((.........)))))))))))))))))))).......... ( -25.90, z-score = -3.32, R) >droSim1.chr3L 11705285 64 - 22553184 UACGUGAUGGCGGCACAUGUCGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCAAGGAG ..(((..(((((((((((((((.((.........)))))))))))))))))))).......... ( -25.90, z-score = -3.32, R) >droSec1.super_0 4511823 64 - 21120651 UACGUGAUGGCGGCACAUGUUGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCAGGGAG ..(((..((((((((((((((................))))))))))))))))).......... ( -23.29, z-score = -2.38, R) >droYak2.chr3L 12381725 64 - 24197627 UACGUGAUGGCAGCACAUGUCGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCGAGGAG ..(((..(((((((((((((((.((.........))))))))))))))))).....)))..... ( -28.30, z-score = -4.19, R) >droEre2.scaffold_4784 12322775 64 - 25762168 UACGUGACGGCAGCACAUGUCGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCAAGGAG ..(((...((((((((((((((.((.........)))))))))))))))).))).......... ( -26.00, z-score = -3.49, R) >droAna3.scaffold_13337 21971410 64 + 23293914 UACGUGCUGGCAGCACAUGUCGCAGCGAAAUUAUCAUGACAUGUGCUGCCACUGGCGCAAGGAC ...(((((((((((((((((((..............))))))))))))))...)))))...... ( -30.84, z-score = -4.22, R) >dp4.chrXR_group6 2683588 63 - 13314419 UACGUGAUGGCAGCACAUGUCGCUGCAAAAUUAUCAUGACAUGUGCUG-CCACAGUGCCAGGAA (((.((.(((((((((((((((.((.........))))))))))))))-))))))))....... ( -27.80, z-score = -3.71, R) >droPer1.super_9 979720 63 - 3637205 UACGUGAUGGCAGCACAUGUCGCUGCAAAAUUAUCAUGACAUGUGCUG-CCACAGUGCCAGGAA (((.((.(((((((((((((((.((.........))))))))))))))-))))))))....... ( -27.80, z-score = -3.71, R) >droWil1.scaffold_180949 2265781 64 + 6375548 UACGUGAUGGCAGCAUAUGUUGCUGCAAAAUUAUCAUGGCAUGUGCUGCCCCCGAUGCUGCGAG ..((.(..(((((((((((((................))))))))))))).))).......... ( -19.99, z-score = -0.64, R) >droMoj3.scaffold_6680 5935054 60 - 24764193 UACGUGCUGGCAGCAUGUGCCGCUGCACAAGCAUCA----GCAGGACGCGAAUGAAGCAUGAAG ..(.((((((..((.(((((....))))).)).)))----))).)..((.......))...... ( -19.90, z-score = -0.53, R) >consensus UACGUGAUGGCAGCACAUGUCGCUGCGAAAUUAUCAUGACAUGUGCUGCCAAUGGAGCAAGGAG ........((((((((((((((..............)))))))))))))).............. (-19.75 = -19.83 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:50 2011