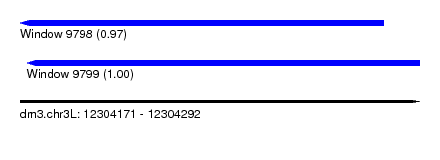
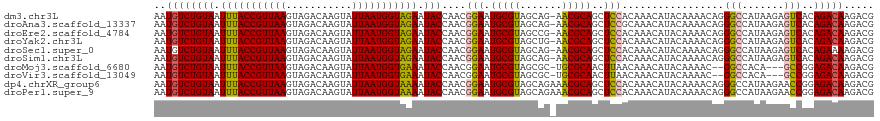
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,304,171 – 12,304,292 |
| Length | 121 |
| Max. P | 0.995573 |

| Location | 12,304,171 – 12,304,281 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.96 |
| Shannon entropy | 0.16165 |
| G+C content | 0.42968 |
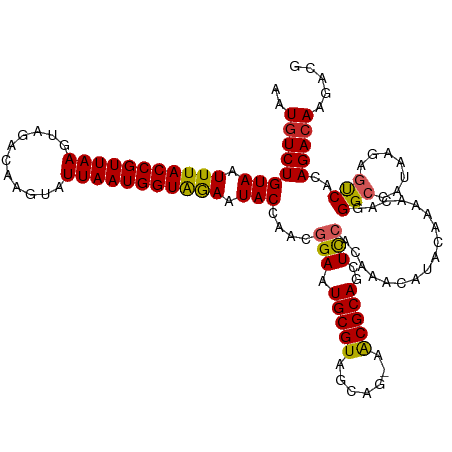
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12304171 110 - 24543557 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........)))))))))))........(((.(((((.....-.)))))..)))...........................(((........)))... ( -22.60, z-score = -2.69, R) >droAna3.scaffold_13337 21948741 110 + 23293914 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCGCAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........))))))))))).......((((.(((((.....-.)))))..))))..........................(((........)))... ( -23.90, z-score = -2.69, R) >droEre2.scaffold_4784 12299531 110 - 25762168 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCCG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........)))))))))))........(((.(((((.....-.)))))..)))...........................(((........)))... ( -22.60, z-score = -2.32, R) >droYak2.chr3L 12357103 110 - 24197627 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCUG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........)))))))))))........(((.(((((.....-.)))))..)))...........................(((........)))... ( -22.60, z-score = -1.98, R) >droSec1.super_0 4488912 110 - 21120651 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGAAAAGACGAG (((((((((((...........)))))))))))........(((.(((((.....-.)))))..)))...........................(((........)))... ( -22.60, z-score = -2.82, R) >droSim1.chr3L 11682867 110 - 22553184 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........)))))))))))........(((.(((((.....-.)))))..)))...........................(((........)))... ( -22.60, z-score = -2.69, R) >droMoj3.scaffold_6680 5915097 105 - 24764193 UUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGC-UGCGCAACUUAACAAACAUACAAAAC--GGCCACA---GCCGGAGACAAGACGAG (((((((((((...........))))))))))).......((...((((((....-)))))).(((................--(((....---)))(....)))).)).. ( -24.60, z-score = -1.96, R) >droVir3.scaffold_13049 7661652 105 - 25233164 UUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGC-UGCGCAACUUAACAAACAUACAAAAC--GGCCACA---GCCGGAGACAAGACGAG (((((((((((...........))))))))))).......((...((((((....-)))))).(((................--(((....---)))(....)))).)).. ( -24.60, z-score = -1.96, R) >dp4.chrXR_group6 2656939 111 - 13314419 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCAGAAACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAACCGGAGACAAGACGAA (((((((((((...........)))))))))))...((...(((.(((((.......)))))..)))................((..(....)..))))............ ( -23.80, z-score = -2.98, R) >droPer1.super_9 953351 111 - 3637205 UUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCAGAAACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAACCGGAGACAAGACGAA (((((((((((...........)))))))))))...((...(((.(((((.......)))))..)))................((..(....)..))))............ ( -23.80, z-score = -2.98, R) >consensus UUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG_AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACGAG (((((((((((...........)))))))))))........(((.(((((.......)))))..))).................(((.......))).............. (-19.58 = -19.02 + -0.56)
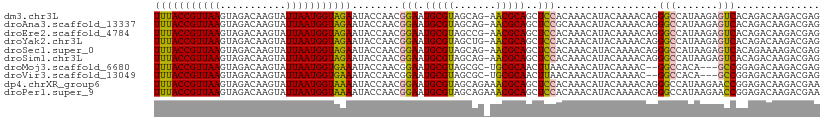
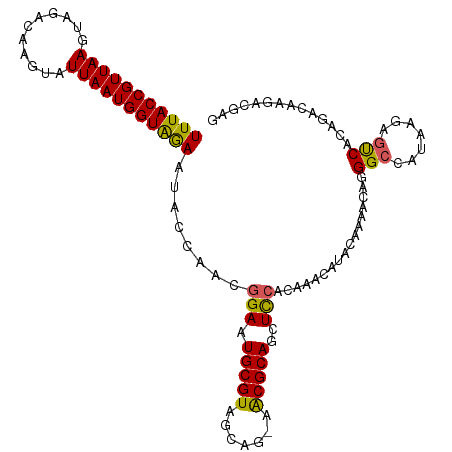
| Location | 12,304,173 – 12,304,292 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Shannon entropy | 0.14351 |
| G+C content | 0.41553 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -26.29 |
| Energy contribution | -25.83 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12304173 119 - 24543557 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..(((((((..(((((((((((...........)))))))))))........(((.(((((.....-.)))))..))).................((((....).))))))))))..... ( -30.80, z-score = -3.83, R) >droAna3.scaffold_13337 21948743 119 + 23293914 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCGCAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..(((((((..(((((((((((...........))))))))))).......((((.(((((.....-.)))))..))))................((((....).))))))))))..... ( -32.10, z-score = -3.86, R) >droEre2.scaffold_4784 12299533 119 - 25762168 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCCG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..(((((((..(((((((((((...........)))))))))))........(((.(((((.....-.)))))..))).................((((....).))))))))))..... ( -30.80, z-score = -3.52, R) >droYak2.chr3L 12357105 119 - 24197627 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCUG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..(((((((..(((((((((((...........)))))))))))........(((.(((((.....-.)))))..))).................((((....).))))))))))..... ( -30.80, z-score = -3.28, R) >droSec1.super_0 4488914 119 - 21120651 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGAAAAGACG ...(((((((.(((((((((((...........))))))))))).)))....(((.(((((.....-.)))))..)))................)))).......(((........))). ( -27.70, z-score = -2.92, R) >droSim1.chr3L 11682869 119 - 22553184 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG-AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..(((((((..(((((((((((...........)))))))))))........(((.(((((.....-.)))))..))).................((((....).))))))))))..... ( -30.80, z-score = -3.83, R) >droMoj3.scaffold_6680 5915099 114 - 24764193 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGC-UGCGCAACUUAACAAACAUACAAAAC--GGCCACA---GCCGGAGACAAGACG ..((((((((.(((((((((((...........))))))))))).)))........((((((....-))))))...................(--(((....---)))).)))))..... ( -30.60, z-score = -2.55, R) >droVir3.scaffold_13049 7661654 114 - 25233164 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGC-UGCGCAACUUAACAAACAUACAAAAC--GGCCACA---GCCGGAGACAAGACG ..((((((((.(((((((((((...........))))))))))).)))........((((((....-))))))...................(--(((....---)))).)))))..... ( -30.60, z-score = -2.55, R) >dp4.chrXR_group6 2656941 120 - 13314419 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCAGAAACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAACCGGAGACAAGACG ..(((((....(((((((((((...........)))))))))))...((...(((.(((((.......)))))..)))................((..(....)..)))))))))..... ( -29.50, z-score = -3.29, R) >droPer1.super_9 953353 120 - 3637205 AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCAGAAACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAACCGGAGACAAGACG ..(((((....(((((((((((...........)))))))))))...((...(((.(((((.......)))))..)))................((..(....)..)))))))))..... ( -29.50, z-score = -3.29, R) >consensus AAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAG_AACGCAGCUCCACAAACAUACAAAACAGGGCCAUAAGAGUCACAGACAAGACG ..((((((((.(((((((((((...........))))))))))).)))....(((.(((((.......)))))..))).................(((.......)))..)))))..... (-26.29 = -25.83 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:48 2011