| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,294,173 – 12,294,275 |
| Length | 102 |
| Max. P | 0.964077 |

| Location | 12,294,173 – 12,294,275 |
|---|---|
| Length | 102 |
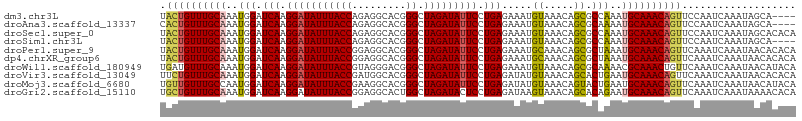
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Shannon entropy | 0.19351 |
| G+C content | 0.39134 |
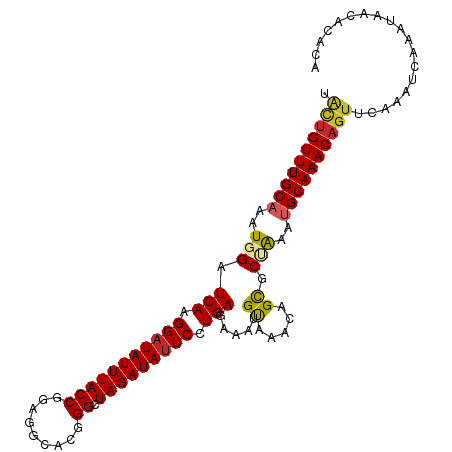
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12294173 102 + 24543557 UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCCAAAUGCAAACAGUUCCAAUCAAAUAGCA---- .((((((((((..(((.(((.(((((((((((.(.....).)).))))))))).))).....((.....)).)))..))))))))))...............---- ( -26.50, z-score = -2.30, R) >droAna3.scaffold_13337 21940106 102 - 23293914 CACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCAAAAUGCAAACAGUUCCAAUCAAAUAGCA---- .((((((((((......(((.(((((((((((.(.....).)).))))))))).)))(...((....))...)....))))))))))...............---- ( -24.00, z-score = -1.62, R) >droSec1.super_0 4478810 106 + 21120651 UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCCAAAUGCAAACAGUUCCAAUCAAAUAGCACACA .((((((((((..(((.(((.(((((((((((.(.....).)).))))))))).))).....((.....)).)))..))))))))))................... ( -26.50, z-score = -2.24, R) >droSim1.chr3L 11672357 102 + 22553184 UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCCAAAUGCAAACAGUUCCAAUCAAAUAGCA---- .((((((((((..(((.(((.(((((((((((.(.....).)).))))))))).))).....((.....)).)))..))))))))))...............---- ( -26.50, z-score = -2.30, R) >droPer1.super_9 941286 106 + 3637205 UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACGGGCUAGAUAUUCCUGAGAAAUGCAAACAGCGCUAAAUGCAAACAGUUCAAAUCAAAUAACACACA .((((((((((..(((.(((.((((((((((((......)))..))))))))).))).....((.....)).)))..))))))))))................... ( -28.70, z-score = -3.59, R) >dp4.chrXR_group6 2646591 106 + 13314419 UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACGGGCUAGAUAUUCCUGAGAAAUGCAAACAGCGCUAAAUGCAAACAGUUCAAAUCAAAUAACACACA .((((((((((..(((.(((.((((((((((((......)))..))))))))).))).....((.....)).)))..))))))))))................... ( -28.70, z-score = -3.59, R) >droWil1.scaffold_180949 2219331 106 - 6375548 UGAUGUUUGCAAAUGGAUCAAGGAUAUUUACCGUAGGGACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCAAAACGCAAACUGUUCAAAUCAAAUAACAUACA (((((((((((......(((.(((((((((((((....))))..))))))))).)))....))))))).(((.....)))...........))))........... ( -25.00, z-score = -2.24, R) >droVir3.scaffold_13049 7651584 106 + 25233164 UUCUGUUUGCAAAUGGAUCAAGGAUAUUUACCGAUGGCACGGGCUAGAUAUUCCUGAGAUAUGUAAACAGCACUGAAUGCAAACAGUUCAAAUCAAAUAACACACA ..(((((((((.(((..(((.((((((((((((......)))..))))))))).)))..))).....((....))..))))))))).................... ( -25.10, z-score = -2.07, R) >droMoj3.scaffold_6680 5905222 106 + 24764193 UGUUGUUUGCCAAUGGAUCAAGGAUAUUUACCGAAGGCACGGGCUAGAUAUUCCUGAGAUAUGUAAACAGUACUGAAUGCAAACAGUUCAAAUCAAAUAACAUACA (((((((((...(((..(((.((((((((((((......)))..))))))))).)))..))).......(.((((........)))).)....))))))))).... ( -23.50, z-score = -1.30, R) >droGri2.scaffold_15110 19340918 106 - 24565398 UGCUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACUGGCUAGAUACUCCUGAGAUAAGUAAACAGCACAGAAUGCAAACAGUUCAAAUCAAAUAAAACACA ((((((((((...((..(((.(((((((((((((.....)))).)))))).))))))..)).))))))))))..((((.......))))................. ( -28.10, z-score = -3.11, R) >consensus UACUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCUAAAUGCAAACAGUUCAAAUCAAAUAACACACA .((((((((((..(((.(((.(((((((((((.........)).))))))))).))).....((.....)).)))..))))))))))................... (-21.46 = -21.52 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:45 2011