| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,277,858 – 12,277,965 |
| Length | 107 |
| Max. P | 0.924915 |

| Location | 12,277,858 – 12,277,965 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.05 |
| Shannon entropy | 0.57205 |
| G+C content | 0.50041 |
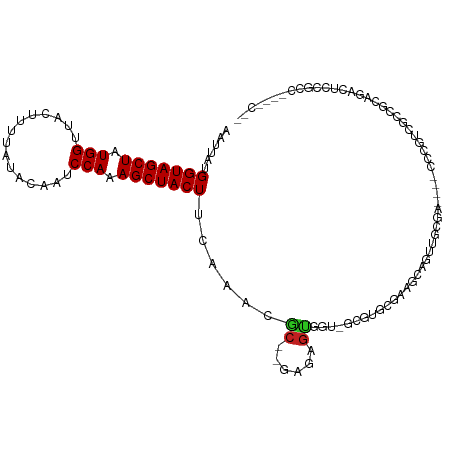
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.51 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
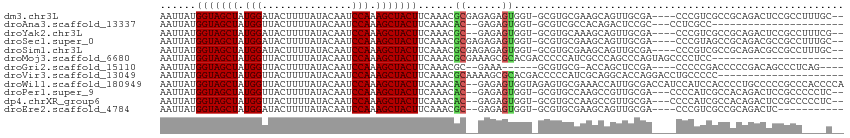
>dm3.chr3L 12277858 107 - 24543557 AAUUAUGGUAGCUAUGGAUACUUUUAUACAAUCCAAAGCUACUUCAAACGCGAGAGAGUGGU-GCGUGCGAAGCAGUUGCGA----CCCGUCGCCGCAGACUCCGCCUUUGC-- ......(((((((.(((((...........))))).))))))).((((.(((.(((.(((((-(((.(.(..((....))..----)))).))))))...)))))).)))).-- ( -36.40, z-score = -2.50, R) >droAna3.scaffold_13337 21926507 86 + 23293914 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACAC--GAGAGUGGU-GCGUCGCCACAGACUCCGC---CCUCGCC---------------------- ......(((((((.(((...............))).))))))).......(--(((.((((.-..(((......))).))))---.))))..---------------------- ( -25.36, z-score = -2.97, R) >droYak2.chr3L 12329797 105 - 24197627 AAUUAUGGUAGCUAUGGAUACUUUUAUACAAUCCAAAGCUACUUCAAACGC--GAGAGUGGU-GCGUGCAAAGCAGUUGCGA----CCCGUCGCCGCAGACUCCGCCUUUCG-- ......(((((((.(((((...........))))).)))))))......((--(.((((..(-(((.((...((....))((----....)))))))).)))))))......-- ( -33.50, z-score = -2.34, R) >droSec1.super_0 4462593 107 - 21120651 AAUUAUGGUAGCUAUGGAUACUUUUAUACAAUCCAAAGCUACUUCAAACGCGAGAGAGUGGU-GCGUGCGAAGCAGUUGCGA----CCCGUAGCCGCAGACGCCGCCUUUGC-- ......(((((((.(((((...........))))).)))))))......((.((((.(((((-(..((((.....((((((.----..))))))))))..))))))))))))-- ( -36.70, z-score = -2.48, R) >droSim1.chr3L 11656247 107 - 22553184 AAUUAUGGUAGCUAUGGAUACUUUUAUACAAUCCAAAGCUACUUCAAACGCGAGAGAGUGGU-GCGUGCGAAGCAGUUGCGA----CCCGUCGCCGCAGACGCCGCCUUUGC-- ......(((((((.(((((...........))))).)))))))......((.((((.(((((-(..((((........((((----....))))))))..))))))))))))-- ( -36.80, z-score = -2.28, R) >droMoj3.scaffold_6680 5885691 92 - 24764193 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACGCGAAAGCGCACGACCCCCAUCGCCCAGCCCAGUAGCCCCUCC---------------------- ......((((((....))))))...............((((((.....(((....)))..(((......)))........))))))......---------------------- ( -18.80, z-score = -1.88, R) >droGri2.scaffold_15110 19320570 97 + 24565398 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACGC--GAAA------GCGUGCG-ACCAGCUCCGA----CCCCCGACCCCCGACAGCCCUCAG---- ......(((((((.(((...............))).)))))))((..((((--....------))))..)-)...(((.((.----...........))..)))......---- ( -20.86, z-score = -2.04, R) >droVir3.scaffold_13049 7632451 93 - 25233164 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACGCAAAAGCGCACGACCCCCAUCGCAGGCACCAGGACCUGCCCCC--------------------- ......(((((((.(((...............))).))))))).....(((....))).............(((((........)))))....--------------------- ( -21.36, z-score = -1.55, R) >droWil1.scaffold_180949 2189774 112 + 6375548 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACAC--GAGAGUGGUAGAGUGCGAAACCAUUGCGACCAUCCAUCCACCCCUGCCCCCGCCCACCCCA ....(((((.((.((((((..................((.((((....(((--....)))...))))))..)))))).)).)))))............................ ( -21.35, z-score = -0.89, R) >droPer1.super_9 922039 106 - 3637205 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACAC--GAGAGUGGU-GCGUGCCAAGCCGUUGCGA---CCCCAUCGCCACAGACUCCGCCCCCUC-- ......(((((((.(((...............))).))))))).......(--(.(((((((-....))).....((.((((---.....)))).))..)))))).......-- ( -25.16, z-score = -1.11, R) >dp4.chrXR_group6 2626909 106 - 13314419 AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACAC--GAGAGUGGU-GCGUGCCAAGCCGUUGCGA---CCCCAUCGCCACAGACUCCGCCCCCUC-- ......(((((((.(((...............))).))))))).......(--(.(((((((-....))).....((.((((---.....)))).))..)))))).......-- ( -25.16, z-score = -1.11, R) >droEre2.scaffold_4784 12272121 96 - 25762168 AAUUAUGGUAGCUAUGGAUACUUUUAUACAAUCCAAAGCUACUUCAAACGC--GAGAGUGGU-GCGUGCGAAGCAGUUGCGA----CCCGUCGCCGCAGACUC----------- ......(((((((.(((((...........))))).))))))).....(((--....))).(-(((.((((.((....))..----....)))))))).....----------- ( -30.90, z-score = -2.09, R) >consensus AAUUAUGGUAGCUAUGGUUACUUUUAUACAAUCCAAAGCUACUUCAAACGC__GAGAGUGGU_GCGUGCGAAGCAGUUGCGA____CCCGUCGCCGCAGACUCCGCC____C__ ......(((((((.(((...............))).)))))))....................................................................... (-11.51 = -11.51 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:42 2011