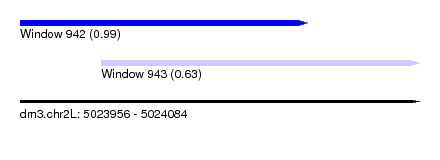
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,023,956 – 5,024,084 |
| Length | 128 |
| Max. P | 0.985591 |

| Location | 5,023,956 – 5,024,048 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Shannon entropy | 0.16261 |
| G+C content | 0.62992 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.09 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
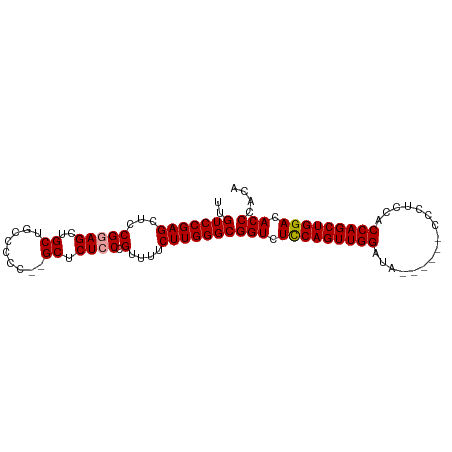
>dm3.chr2L 5023956 92 + 23011544 UUGUCCGAGCUCCG-AGCUGCUGCCCCC--GCUCUUC-GUUUUCUUGGGCGGUCUUCAGUUGGAUCGAAAUCCCCUCCACCAGCUGGACACCCAUC ..(((((((...((-((..((.......--))..)))-)....)))))))(((.(((((((((...((.......))..))))))))).))).... ( -29.60, z-score = -1.66, R) >droSec1.super_5 3110367 88 + 5866729 UUGUCCGAGCUCCGGAGCUGCUGCCCCC--GCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAUA------CCCUCCUCCAGCUGGACACCCACA ..(((((((..(.((((..((.......--)).)))).)....)))))))(((.((((((((((..------......)))))))))).))).... ( -36.70, z-score = -3.54, R) >droSim1.chr2L 4875717 90 + 22036055 UUGUCCGAGCUCCGGAGCUGCUGCCCCCCCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGACA------CCCUCCACCAGCUGGACACCCACA ..(((((((..(.((((..((.........)).)))).)....)))))))(((.(((((((((...------.......))))))))).))).... ( -33.70, z-score = -2.35, R) >consensus UUGUCCGAGCUCCGGAGCUGCUGCCCCC__GCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAUA______CCCUCCACCAGCUGGACACCCACA ..(((((((..(.((((..((.........)).)))).)....)))))))(((.(((((((((................))))))))).))).... (-29.98 = -30.09 + 0.11)

| Location | 5,023,982 – 5,024,084 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Shannon entropy | 0.13373 |
| G+C content | 0.64918 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -34.53 |
| Energy contribution | -34.09 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
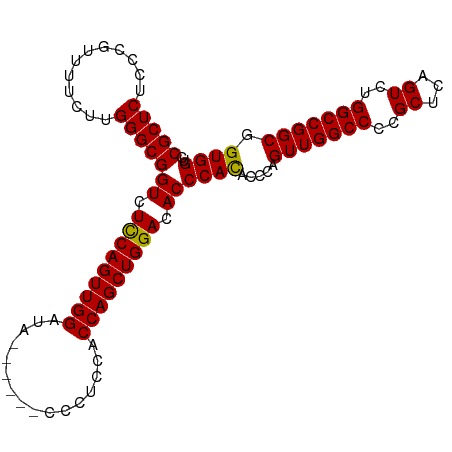
>dm3.chr2L 5023982 102 + 23011544 CGCUCUUC-GUUUUCUUGGGCGGUCUUCAGUUGGAUCGAAAUCCCCUCCACCAGCUGGACACCCAUCACCAGUUGGCCCCGCUCAGUCUGGCCGGCGGUGGUC .((((...-........))))(((.(((((((((...((.......))..))))))))).))).((((((.(((((((..((...))..))))))))))))). ( -37.60, z-score = -1.33, R) >droSec1.super_5 3110394 97 + 5866729 CGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAUA------CCCUCCUCCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGGCCGGCGGUGGUC .((((............))))(((.((((((((((..------......)))))))))).)))..((((..(((((((..((...))..)))))))))))... ( -40.40, z-score = -1.97, R) >droSim1.chr2L 4875746 97 + 22036055 CGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGACA------CCCUCCACCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGGCCGGCGGUGGUC .((((............))))(((.(((((((((...------.......))))))))).)))..((((..(((((((..((...))..)))))))))))... ( -37.90, z-score = -0.94, R) >consensus CGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAUA______CCCUCCACCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGGCCGGCGGUGGUC (((((............)))))((.(((((((((................))))))))).))((((.....(((((((..((...))..))))))).)))).. (-34.53 = -34.09 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:48 2011