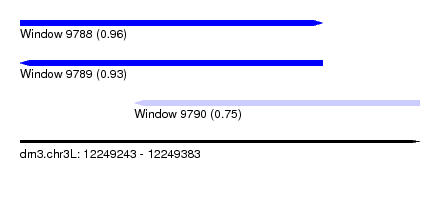
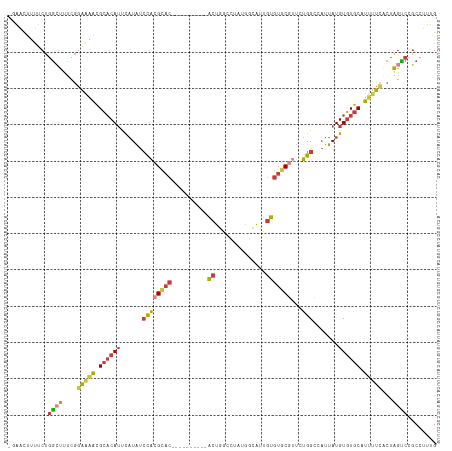
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,249,243 – 12,249,383 |
| Length | 140 |
| Max. P | 0.963065 |

| Location | 12,249,243 – 12,249,349 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.53659 |
| G+C content | 0.48563 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -13.00 |
| Energy contribution | -15.78 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
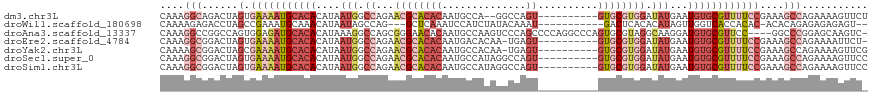
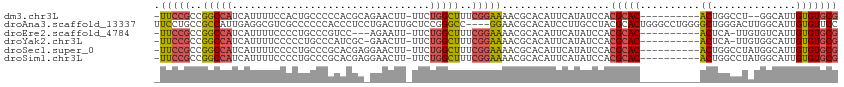
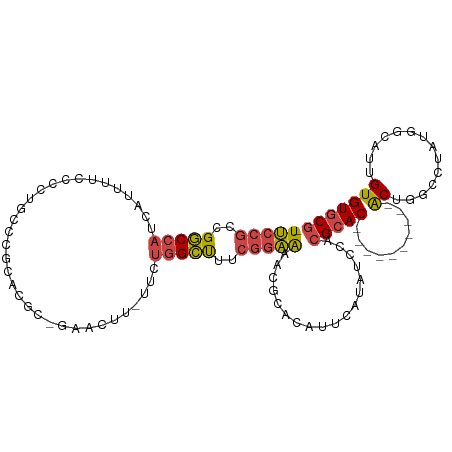
>dm3.chr3L 12249243 106 + 24543557 CAAAGGCAGACUAGUGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGCCA--GGCCAGU----------GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAGUUCU ....(((......(.(((((((((((((((....((...((((((((...((..--.))..))----------)))))))).))))..))))))))))))....)))........... ( -33.30, z-score = -2.47, R) >droWil1.scaffold_180698 2689570 101 + 11422946 CAAAAGAGACCUAGCCGAAAUGCAAACAUAAUAGCCAG---GCUCAAAUCCAUCUAUACAAAU-----------GACUCACACAUAGUUGGUCACCACAC-ACACAGAGAGAGAGU-- .....(((.(((.((....(((....)))....)).))---))))...((..(((.......(-----------((((.((.....)).)))))......-....)))..))....-- ( -15.43, z-score = -0.92, R) >droAna3.scaffold_13337 21899132 113 - 23293914 CAAAGGCCGGCCAGUGGAGAUGCACACAUAAAGGCCAGCGGGAACACAAUGCCAAGUCCCAGCCCCAGGCCCAGUGCGUAGGCAAGGAUGUGCGUUCC----GGCCCGGAGCAAGUC- ....((((((........((((((((......((((.((((((.............)))).))....))))(..(((....)))..).))))))))))----))))...........- ( -36.42, z-score = 0.56, R) >droEre2.scaffold_4784 12240163 106 + 25762168 CAAAGGCGGACUAGUGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGACACAA-UGAGU----------GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAUUCU- ....(((......(.(((((((((((((((....((...((((((((.((......)-)..))----------)))))))).))))..))))))))))))....)))..........- ( -34.30, z-score = -3.59, R) >droYak2.chr3L 12300334 107 + 24197627 CAAAAGCGGACUAGCGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGCCACAA-UGAGU----------GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAGUUCG ......((((((...(((((((((((((((....((...((((((((.((......)-)..))----------)))))))).))))..)))))))))))(....).......)))))) ( -34.50, z-score = -3.22, R) >droSec1.super_0 4434179 108 + 21120651 CAAAGGCGGACUAGUGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGCCAUAGGCCAGU----------GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAGUUCC ....(((......(.(((((((((((((((....((...((((((((...(((...)))..))----------)))))))).))))..))))))))))))....)))........... ( -37.70, z-score = -3.68, R) >droSim1.chr3L 11625202 108 + 22553184 CAAAGGCGGACUAGUGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGCCAUAGGCCAGU----------GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAGUUCC ....(((......(.(((((((((((((((....((...((((((((...(((...)))..))----------)))))))).))))..))))))))))))....)))........... ( -37.70, z-score = -3.68, R) >consensus CAAAGGCGGACUAGUGAAAAUGCACACAUAAUGGCCAGAACGCACACAAUGCCAUAGGCCAGU__________GUGCGUGGAUAUGAAUGUGCGUUUUCCGAAAGCCAGAAAAGUUC_ ....(((......(.(((((((((((....(((.((...((((((............................)))))))).)))...))))))))))))....)))........... (-13.00 = -15.78 + 2.78)

| Location | 12,249,243 – 12,249,349 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.53659 |
| G+C content | 0.48563 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
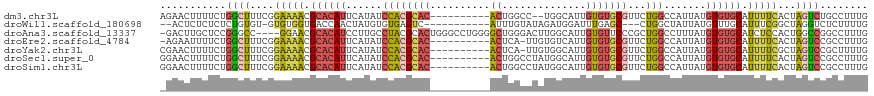
>dm3.chr3L 12249243 106 - 24543557 AGAACUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCC--UGGCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCACUAGUCUGCCUUUG ...........(((...(.(((((.((((((......((((((((----------((..((.--..))...))))))))...)).......)))))).)))))....)...))).... ( -31.52, z-score = -2.20, R) >droWil1.scaffold_180698 2689570 101 - 11422946 --ACUCUCUCUCUGUGU-GUGUGGUGACCAACUAUGUGUGAGUC-----------AUUUGUAUAGAUGGAUUUGAGC---CUGGCUAUUAUGUUUGCAUUUCGGCUAGGUCUCUUUUG --........(((((.(-(((..(((((..((.....))..)))-----------).)..)))).)))))...((((---((((((...(((....)))...))))))).)))..... ( -30.50, z-score = -2.94, R) >droAna3.scaffold_13337 21899132 113 + 23293914 -GACUUGCUCCGGGCC----GGAACGCACAUCCUUGCCUACGCACUGGGCCUGGGGCUGGGACUUGGCAUUGUGUUCCCGCUGGCCUUUAUGUGUGCAUCUCCACUGGCCGGCCUUUG -.....((((((((((----(((.......))).(((....)))...)))))))))).((((....((.....))))))(((((((.....(((........))).)))))))..... ( -44.60, z-score = -1.00, R) >droEre2.scaffold_4784 12240163 106 - 25762168 -AGAAUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUCA-UUGUGUCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCACUAGUCCGCCUUUG -..........(((...(.(((((.((((((......((((((((----------((...-..........))))))))...)).......)))))).)))))....)...))).... ( -28.84, z-score = -2.32, R) >droYak2.chr3L 12300334 107 - 24197627 CGAACUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUCA-UUGUGGCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCGCUAGUCCGCUUUUG ...........((((..((((((.(((((((......((((((((----------((...-..........))))))))...)).....)))))))..))))))..))))........ ( -29.62, z-score = -1.75, R) >droSec1.super_0 4434179 108 - 21120651 GGAACUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCCUAUGGCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCACUAGUCCGCCUUUG (((..(((((((.....))))))).((((((......((((((((----------((..(((...)))...))))))))...)).......))))))...........)))....... ( -34.22, z-score = -2.85, R) >droSim1.chr3L 11625202 108 - 22553184 GGAACUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCCUAUGGCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCACUAGUCCGCCUUUG (((..(((((((.....))))))).((((((......((((((((----------((..(((...)))...))))))))...)).......))))))...........)))....... ( -34.22, z-score = -2.85, R) >consensus _GAACUUUUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC__________ACUGGCCUAUGGCAUUGUGUGCGUUCUGGCCAUUAUGUGUGCAUUUUCACUAGUCCGCCUUUG ...........((((....(((((.((((((......((((((((............................)))))...))).......)))))).)))))...))))........ (-12.90 = -13.24 + 0.34)

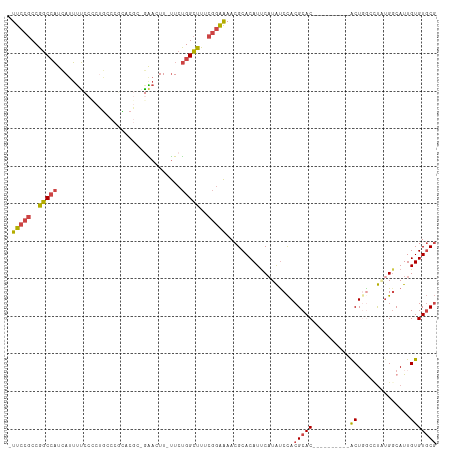
| Location | 12,249,283 – 12,249,383 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.45276 |
| G+C content | 0.55865 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.69 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12249283 100 - 24543557 -UUCCGCCGGCCAUCAUUUUCCACUGCCCCCACGCAGAACUU-UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCCU--GGCAUUGUGUGCG -(((((..(((((..........((((......)))).....-...)))))..)))))..(((((((......(((.((..----------....)).)--))....))))))) ( -30.87, z-score = -2.86, R) >droAna3.scaffold_13337 21899172 110 + 23293914 UUCCUGCCGCCAUUGAGGCGUCGCCCCCCACCCUCCUGACUUGCUCCGGGCC----GGAACGCACAUCCUUGCCUACGCACUGGGCCUGGGGCUGGGACUUGGCAUUGUGUUCC ....(((((((.....))).......((((..(....)....((((((((((----(((.......))).(((....)))...))))))))))))))....))))......... ( -38.80, z-score = 0.52, R) >droEre2.scaffold_4784 12240203 98 - 25762168 -UUCCGCCGGCCAUCAUUUUCCCCUGCCCGUCC---AGAAUU-UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUCA-UUGUGUCAUUGUGUGCG -(((((..(((((..(..(((............---.)))..-)..)))))..)))))..(((((((........(((((.----------.....-.)))))....))))))) ( -23.92, z-score = -2.00, R) >droYak2.chr3L 12300374 100 - 24197627 -UUCCGCCGGCCAUCAUUUUCCCCCUGCCCAUCGC-GAACUU-UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUCA-UUGUGGCAUUGUGUGCG -(((((..(((((.............((.....))-((....-.)))))))..)))))..(((((((......(((((...----------.....-.)))))....))))))) ( -26.30, z-score = -1.99, R) >droSec1.super_0 4434219 102 - 21120651 -UUCCGCCGGCCAUCAUUUUCCCCUGCCCGCACGAGGAACUU-UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCCUAUGGCAUUGUGUGCG -(((((..(((((.....(((((.((....)).).))))...-...)))))..)))))..................(((((----------((..(((...)))...))))))) ( -33.10, z-score = -2.71, R) >droSim1.chr3L 11625242 102 - 22553184 -UUCCGCCGGCCAUCAUUUUCCCCUGCCCGCACGAGGAACUU-UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC----------ACUGGCCUAUGGCAUUGUGUGCG -(((((..(((((.....(((((.((....)).).))))...-...)))))..)))))..................(((((----------((..(((...)))...))))))) ( -33.10, z-score = -2.71, R) >consensus _UUCCGCCGGCCAUCAUUUUCCCCUGCCCGCACGC_GAACUU_UUCUGGCUUUCGGAAAACGCACAUUCAUAUCCACGCAC__________ACUGGCCUAUGGCAUUGUGUGCG .(((((..(((((.................................)))))..)))))..(((((((......(((........................)))....))))))) (-14.15 = -14.69 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:40 2011