| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,215,953 – 12,216,043 |
| Length | 90 |
| Max. P | 0.924073 |

| Location | 12,215,953 – 12,216,043 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.67503 |
| G+C content | 0.43952 |
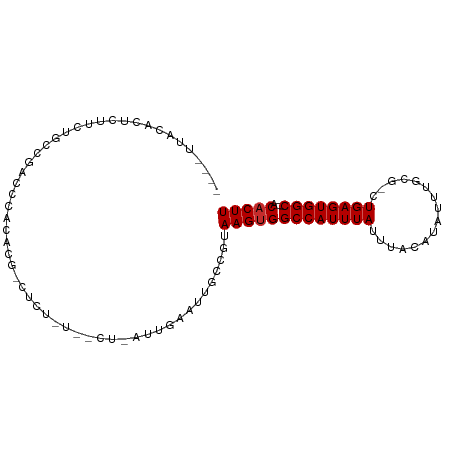
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.924073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
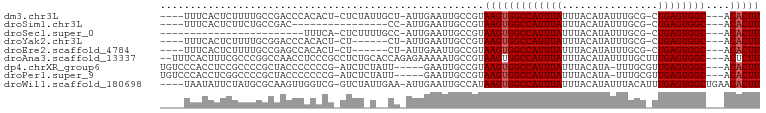
>dm3.chr3L 12215953 90 - 24543557 ----UUUCACUCUUUUGCCGACCCACACU-CUCUAUUGCU-AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG-CUGAGUGGC---ACACUU ----..((((((.............((((-...(((.((.-........)).)))))))((((..(((....)))..)).)-).)))))).---...... ( -14.20, z-score = 0.03, R) >droSim1.chr3L 11590196 75 - 22553184 ----UUUCACUCUUCUGCCGAC----------------CC-AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG-CUGAGUGGC---ACACUU ----..((((((....(((((.----------------..-..(((((.((((....)))).))))).........))).)-).)))))).---...... ( -14.74, z-score = -0.65, R) >droSec1.super_0 4401074 70 - 21120651 ------------------------UUUCA-CUCUUUUGCC-AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG-CUGAGUGGC---ACACUU ------------------------..(((-(((....(((-(.(((((.((((....)))).)))))..........)).)-).)))))).---...... ( -14.10, z-score = -0.70, R) >droYak2.chr3L 12266799 84 - 24197627 ----UUUCACUCUUUUGCGGACCCACACU-CU------CU-AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG-CUGAGUGGC---ACACUU ----..........((((((.....((..-..------..-..)).....))))))(((((((((((....((....))..-.))))))))---.))).. ( -18.40, z-score = -1.57, R) >droEre2.scaffold_4784 12205758 84 - 25762168 ----UUUCACUCUUUUGCCGAGCCACACU-CU------CU-AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG-CUGAGUGGC---ACACUU ----..((((((...(((.(((.....))-).------..-..(((((.((((....)))).)))))...........)))-..)))))).---...... ( -17.10, z-score = -0.90, R) >droAna3.scaffold_13337 21871205 95 + 23293914 --UUUCACUUUCGCCCGGCCAACCUCCCGCCUCUGCACCAGAGAAAAAUGCCGUAAGUGGCCAUUUAUUUACAUAUUUUGCUUUGAGUGGC---ACUCUU --...(((((.((..(((........))).(((((...)))))........)).)))))((((((((....((.....))...))))))))---...... ( -21.70, z-score = -1.34, R) >dp4.chrXR_group6 2556627 90 - 13314419 UGUCCCACCUCCGCCCCGCUACCCCCCCG-AUCUCUAUU-----GAAUUGCCGUAAGUGGCCAUUUAUUUACAUA-UUUGCGUUGAGUGGC---ACACUU (((.(((((..(((.............((-((.((....-----))))))..((((((((....))))))))...-...)))..).)))).---)))... ( -17.10, z-score = -1.27, R) >droPer1.super_9 848731 90 - 3637205 UGUCCCACCUCGGCCCCGCUACCCCCCCG-AUCUCUAUU-----GAAUUGCCGUAAGUGGCCAUUUAUUUACAUA-UUUGCGUUGAGUGGC---ACACUU ..........((((..((.........))-.((......-----))...)))).(((((((((((((....((..-..))...))))))))---.))))) ( -18.20, z-score = -0.66, R) >droWil1.scaffold_180698 2650569 94 - 11422946 ----UAAUAUUCUAUGCGCAAGUUGGUCG-GUCUAUUGAA-AUUGAAUUGCCAUAAGUGGCCAUUUAUUUACAUAUUUACAUUUGAGUGGCUGAACACUU ----.....(((((((.((((.(..((..-.((....)).-))..).))))))))...(((((((((................))))))))))))..... ( -19.09, z-score = -0.51, R) >consensus ____UUACACUCUUCUGCCGACCCACACG_CUCU_U__CU_AUUGAAUUGCCGUAAGUGGCCAUUUAUUUACAUAUUUGCG_CUGAGUGGC___ACACUU ......................................................(((((((((((((................))))))))....))))) ( -9.56 = -9.67 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:33 2011