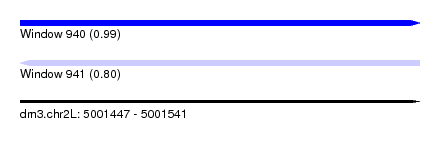
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,001,447 – 5,001,541 |
| Length | 94 |
| Max. P | 0.987522 |

| Location | 5,001,447 – 5,001,541 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.65105 |
| G+C content | 0.43342 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.39 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987522 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
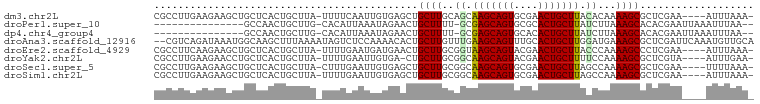
>dm3.chr2L 5001447 94 + 23011544 CGCCUUGAAGAAGCUGCUCACUGCUUA-UUUUCAAUUGUGAGCUGCUUGCAGCAAGCAGUGCGAACUGCUUACACAAAAGCGCUCGAA----AUUUAAA- (((.((((((((((........)))..-)))))))(((((.((((....))))(((((((....))))))).)))))..)))......----.......- ( -29.40, z-score = -1.28, R) >droPer1.super_10 521983 81 + 3432795 ---------------GCCAACUGCUUG-CACAUUAAAUAGAACUGCUUUU-GCGAGCAGUGCGCACUGCUUAUCUUAAAGCACACGAAUUAAAUUUAA-- ---------------((((.(((((((-((...................)-)))))))))).))..(((((......)))))................-- ( -19.71, z-score = -1.52, R) >dp4.chr4_group4 1518183 81 + 6586962 ---------------GCCAACUGCUUG-CACAUUAAAUAGAACUGCUUUU-GCGAGCAGUGCACACUGCUUAUCUUAAAGCACACGAAUUAAAUUUAA-- ---------------((..((((((((-((...................)-)))))))))))....(((((......)))))................-- ( -18.61, z-score = -1.64, R) >droAna3.scaffold_12916 13631216 98 + 16180835 --CGUCAGAUAAAUGGCAAGCUUUAAAAUAGUCUCCAAAACACUGCUUGUUUGAAGCAGUUUGCACUGCUUGGAUGAAAGCGCUCGAUUCAAAUGUUGCA --.((((......))))..(((((......((((...(((((.....))))).(((((((....))))))))))).)))))((..............)). ( -19.84, z-score = 1.12, R) >droEre2.scaffold_4929 5079702 94 + 26641161 CGCCUUCAAGAAGCUGCUCACUGCUUA-UUUUGAAUGAUGAACUGCUUGCGGUAAGCAGUACGAACUGCUUACCCAAAAGCCCUCGAA----AUUUAAA- ..........((((........)))).-.(((((((..(((...((((..((((((((((....))))))))))...))))..)))..----)))))))- ( -27.30, z-score = -3.53, R) >droYak2.chr2L 11799132 93 - 22324452 CGCCUUGAAGAACCUGCUCACUGCUUA-UUUUGAAUUGUGA-CUGCUUGCGGCAAGCAGUACGAACUGCUUUUCCAAAAGCGCUCGUA----AUUUGAA- .((..(((.(......))))..))...-.((..((((((((-.(((((..((.(((((((....)))))))..))..))))).)))))----)))..))- ( -24.50, z-score = -1.37, R) >droSec1.super_5 3087962 94 + 5866729 CGCCUUGAAGAAGCUGCUCACUGCUUA-CUUUGAAUUGUGAGCUGCUUGCGGCAAGCAGUGCGAACUGCUUAGCCAAAAGCGCUCGAA----UUUUAAA- ....(..(((((((........)))).-)))..)....(((((.((((..((((((((((....))))))).)))..)))))))))..----.......- ( -34.80, z-score = -2.46, R) >droSim1.chr2L 4857200 94 + 22036055 CGCCUUGAAGAAGCUGCUCACUGCUUA-UUUUGAAUUGUGAGCUGCUUGCGGCAAGCAGUGCGAACUGCUUAGCCAAAAGCGCUCGAA----AUUUAAA- ..........((((........)))).-.((((((((.(((((.((((..((((((((((....))))))).)))..))))))))).)----)))))))- ( -35.00, z-score = -2.62, R) >consensus CGCCUUGAAGAAGCUGCUCACUGCUUA_UUUUGAAUUGUGAACUGCUUGCGGCAAGCAGUGCGAACUGCUUAGCCAAAAGCGCUCGAA____AUUUAAA_ ............................................((((..((.(((((((....))))))).))...))))................... (-13.32 = -13.39 + 0.06)

| Location | 5,001,447 – 5,001,541 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 66.93 |
| Shannon entropy | 0.65105 |
| G+C content | 0.43342 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -11.74 |
| Energy contribution | -12.16 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799411 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
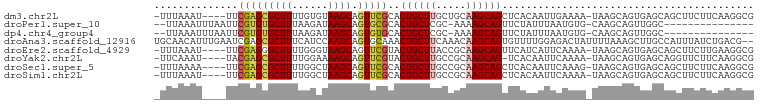
>dm3.chr2L 5001447 94 - 23011544 -UUUAAAU----UUCGAGCGCUUUUGUGUAAGCAGUUCGCACUGCUUGCUGCAAGCAGCUCACAAUUGAAAA-UAAGCAGUGAGCAGCUUCUUCAAGGCG -.......----......((((((.(((((((((((....))))))))).))((((.((((((.........-......)))))).))))....)))))) ( -29.46, z-score = -0.82, R) >droPer1.super_10 521983 81 - 3432795 --UUAAAUUUAAUUCGUGUGCUUUAAGAUAAGCAGUGCGCACUGCUCGC-AAAAGCAGUUCUAUUUAAUGUG-CAAGCAGUUGGC--------------- --......((((((.((.(((((......))))).(((((((((((...-...))))))..........)))-)).)))))))).--------------- ( -16.90, z-score = 0.95, R) >dp4.chr4_group4 1518183 81 - 6586962 --UUAAAUUUAAUUCGUGUGCUUUAAGAUAAGCAGUGUGCACUGCUCGC-AAAAGCAGUUCUAUUUAAUGUG-CAAGCAGUUGGC--------------- --......(((((((.((..(.(((((...((((((....)))))).((-....)).......))))).)..-)).).)))))).--------------- ( -16.40, z-score = 0.95, R) >droAna3.scaffold_12916 13631216 98 - 16180835 UGCAACAUUUGAAUCGAGCGCUUUCAUCCAAGCAGUGCAAACUGCUUCAAACAAGCAGUGUUUUGGAGACUAUUUUAAAGCUUGCCAUUUAUCUGACG-- .....((..((((((((((((((......(((((((....))))))).....))))........(....).........)))))..)))))..))...-- ( -22.30, z-score = -0.46, R) >droEre2.scaffold_4929 5079702 94 - 26641161 -UUUAAAU----UUCGAGGGCUUUUGGGUAAGCAGUUCGUACUGCUUACCGCAAGCAGUUCAUCAUUCAAAA-UAAGCAGUGAGCAGCUUCUUGAAGGCG -......(----((((((((((((((((((((((((....)))))))))).))))..((((((.........-......))))))))).))))))))... ( -32.16, z-score = -3.62, R) >droYak2.chr2L 11799132 93 + 22324452 -UUCAAAU----UACGAGCGCUUUUGGAAAAGCAGUUCGUACUGCUUGCCGCAAGCAG-UCACAAUUCAAAA-UAAGCAGUGAGCAGGUUCUUCAAGGCG -.......----((((((((((((....))))).)))))))(((((((...)))))))-.............-...((..((((.......))))..)). ( -25.20, z-score = -1.23, R) >droSec1.super_5 3087962 94 - 5866729 -UUUAAAA----UUCGAGCGCUUUUGGCUAAGCAGUUCGCACUGCUUGCCGCAAGCAGCUCACAAUUCAAAG-UAAGCAGUGAGCAGCUUCUUCAAGGCG -.......----...((((((((.((((.(((((((....))))))))))).)))).((((((.........-......)))))).)))).......... ( -33.06, z-score = -2.28, R) >droSim1.chr2L 4857200 94 - 22036055 -UUUAAAU----UUCGAGCGCUUUUGGCUAAGCAGUUCGCACUGCUUGCCGCAAGCAGCUCACAAUUCAAAA-UAAGCAGUGAGCAGCUUCUUCAAGGCG -.......----...((((((((.((((.(((((((....))))))))))).)))).((((((.........-......)))))).)))).......... ( -33.06, z-score = -2.62, R) >consensus _UUUAAAU____UUCGAGCGCUUUUGGAUAAGCAGUUCGCACUGCUUGCCGCAAGCAGUUCACAAUUCAAAA_UAAGCAGUGAGCAGCUUCUUCAAGGCG ...................((((.....((((((((....))))))))....))))............................................ (-11.74 = -12.16 + 0.42)
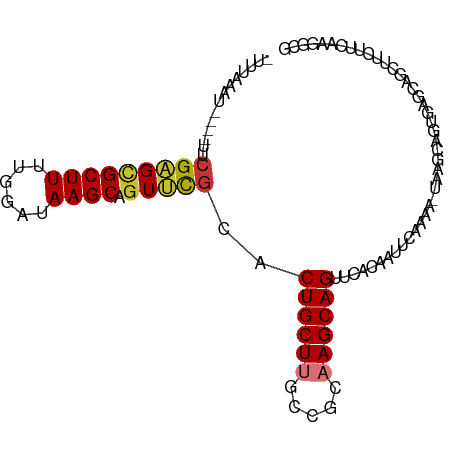
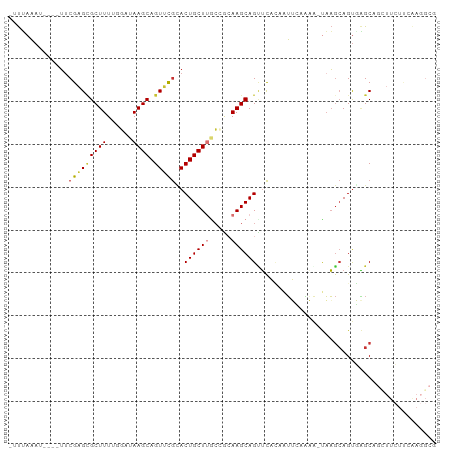
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:47 2011