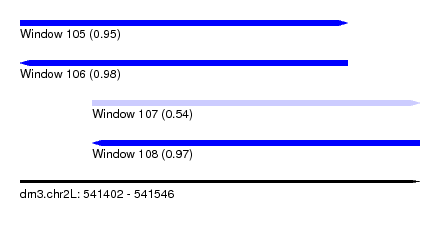
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 541,402 – 541,546 |
| Length | 144 |
| Max. P | 0.982474 |

| Location | 541,402 – 541,520 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.52 |
| Shannon entropy | 0.18705 |
| G+C content | 0.42488 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
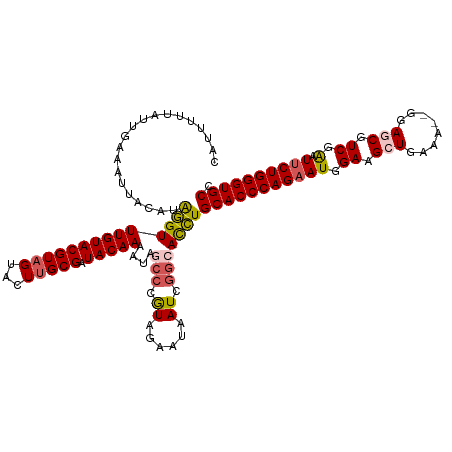
>dm3.chr2L 541402 118 + 23011544 AAUUUUUAUUGAAAUUACAUAGGUUUGUACGUAGUACUCGCGAUACAAAAUAGCCCGUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGAAA--AGAGCGUCGAAUUCUGGGUGCC ((((((....))))))....((((((((((((.......))).)))))....(((.((......)).)))))))(((((((((((.((.(((....--..))).))..))))))))))). ( -36.70, z-score = -3.52, R) >droEre2.scaffold_4929 596449 118 + 26641161 CAUUUUUAUUGAAAUUACAUGUGUUUGUACGUAGUACUUGCGAUACAAAAUAGACCGUAGAAUAAUCAG-AUAUGCACCCAGAAUCGAAGAUGAGA-AUGAGCUUCGGUUACUGGGUGCC ..............((((.....(((((((((((...))))).)))))).......)))).........-....((((((((((((((((......-.....)))))))).)))))))). ( -31.70, z-score = -3.00, R) >droYak2.chr2L 528704 118 + 22324452 CAUUUUUAUUGAAAUUACAUAUGUUUGUACGUAGUACUUGCGAUACAAAAUAGCCCGUAGAACAAUCGG-ACAUGCACCCAGAAUCGAAGAUGGAA-GGGAGCGUCGGCUUCUGGGUGCC .((((((((((.((.(((.(((((....)))))))).)).))))).))))).(.(((.........)))-.)..((((((((((.(...((((...-.....)))).).)))))))))). ( -31.10, z-score = -1.25, R) >droSec1.super_14 523974 119 + 2068291 CAUUUUUAUUGAAAUUACAUAGGUUUGUACGUAGUACUUGCGGUACAAAAUAGCCCAUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGCAA-UGGAGCGUCGAAUUCUGGGUGCC ....................((((((((((((((...)))).))))))....(((.((......)).)))))))(((((((((((.((.(((.(..-.).))).))..))))))))))). ( -36.90, z-score = -2.59, R) >droSim1.chr2L 549899 120 + 22036055 CAUUUUUAUUGAAAUUACAUAGGUUUGUACGUAGUACUUGCGGUACAAAAUAGCCCAUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGCAAGCUGAGCGUCGAAUUCUGGGUGCC ....................((((((((((((((...)))).))))))....(((.((......)).)))))))(((((((((((.((.(((........))).))..))))))))))). ( -36.70, z-score = -2.37, R) >consensus CAUUUUUAUUGAAAUUACAUAGGUUUGUACGUAGUACUUGCGAUACAAAAUAGCCCGUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGAAA__GGAGCGUCGAAUUCUGGGUGCC ....................((((((((((((((...))))).))))))...(((.((......)).))).)))(((((((((((.((.(((........))).)).).)))))))))). (-27.08 = -27.40 + 0.32)

| Location | 541,402 – 541,520 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.52 |
| Shannon entropy | 0.18705 |
| G+C content | 0.42488 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.56 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
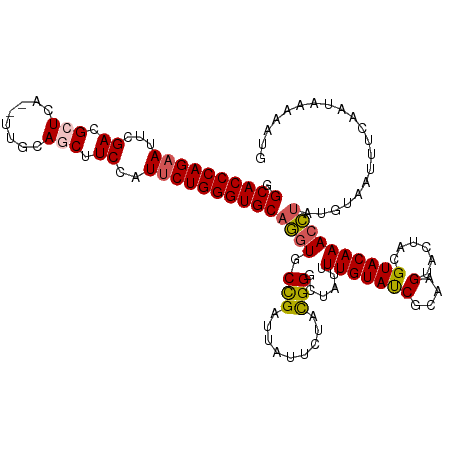
>dm3.chr2L 541402 118 - 23011544 GGCACCCAGAAUUCGACGCUCU--UUUCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUACGGGCUAUUUUGUAUCGCGAGUACUACGUACAAACCUAUGUAAUUUCAAUAAAAAUU .(((((((((((..((.(((..--....))).)).))))))))))).(((((.((.((..(....)..)).)).)))))..(((...((((((......))))))..))).......... ( -35.00, z-score = -3.53, R) >droEre2.scaffold_4929 596449 118 - 26641161 GGCACCCAGUAACCGAAGCUCAU-UCUCAUCUUCGAUUCUGGGUGCAUAU-CUGAUUAUUCUACGGUCUAUUUUGUAUCGCAAGUACUACGUACAAACACAUGUAAUUUCAAUAAAAAUG .((((((((.((.(((((.....-......))))).))))))))))....-..........((((......((((((((....)......)))))))....))))............... ( -25.30, z-score = -2.51, R) >droYak2.chr2L 528704 118 - 22324452 GGCACCCAGAAGCCGACGCUCCC-UUCCAUCUUCGAUUCUGGGUGCAUGU-CCGAUUGUUCUACGGGCUAUUUUGUAUCGCAAGUACUACGUACAAACAUAUGUAAUUUCAAUAAAAAUG .((((((((((..(((.......-........))).))))))))))..((-(((.........)))))(((((((((.(....))))((((((......)))))).........)))))) ( -31.46, z-score = -2.56, R) >droSec1.super_14 523974 119 - 2068291 GGCACCCAGAAUUCGACGCUCCA-UUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUAUGGGCUAUUUUGUACCGCAAGUACUACGUACAAACCUAUGUAAUUUCAAUAAAAAUG .(((((((((((..((.(((...-....))).)).)))))))))))(((((((.((......)).)))....(((((((....)......)))))))))).................... ( -36.70, z-score = -3.41, R) >droSim1.chr2L 549899 120 - 22036055 GGCACCCAGAAUUCGACGCUCAGCUUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUAUGGGCUAUUUUGUACCGCAAGUACUACGUACAAACCUAUGUAAUUUCAAUAAAAAUG .(((((((((((..((.((((.....).))).)).)))))))))))(((((((.((......)).)))....(((((((....)......)))))))))).................... ( -37.30, z-score = -3.23, R) >consensus GGCACCCAGAAUUCGACGCUCA__UUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUACGGGCUAUUUUGUAUCGCAAGUACUACGUACAAACCUAUGUAAUUUCAAUAAAAAUG .((((((((((...((.(((........))).))..))))))))))((((.(((.........)))......(((((((....)......)))))))))).................... (-24.68 = -24.56 + -0.12)

| Location | 541,428 – 541,546 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.22608 |
| G+C content | 0.51278 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -28.64 |
| Energy contribution | -30.00 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 541428 118 + 23011544 GUACGUAGUACUCGCGAUACAAAAUAGCCCGUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGAAA--AGAGCGUCGAAUUCUGGGUGCCACGUGGCCACUUUGGGCCUCAUCGAA ((((...))))...((((........(((.((......)).)))....(((((((((((.((.(((....--..))).))..)))))))))))...(.((((......)))).))))).. ( -40.40, z-score = -2.45, R) >droEre2.scaffold_4929 596475 118 + 26641161 GUACGUAGUACUUGCGAUACAAAAUAGACCGUAGAAUAAUCAG-AUAUGCACCCAGAAUCGAAGAUGAGA-AUGAGCUUCGGUUACUGGGUGCCACGUAGCCACUUUGGGGCUCUUCAAA .(((((.....(((((.............))))).........-....((((((((((((((((......-.....)))))))).)))))))).)))))(((.(....))))........ ( -34.12, z-score = -1.99, R) >droYak2.chr2L 528730 118 + 22324452 GUACGUAGUACUUGCGAUACAAAAUAGCCCGUAGAACAAUCGG-ACAUGCACCCAGAAUCGAAGAUGGAA-GGGAGCGUCGGCUUCUGGGUGCCACGUAGCCACUUUGGGGCUCAUCAUA ((((((((...))))).))).....(((((...((....))((-..((((((((((((.(...((((...-.....)))).).))))))))))...))..))......)))))....... ( -34.30, z-score = -0.25, R) >droSec1.super_14 524000 117 + 2068291 GUACGUAGUACUUGCGGUACAAAAUAGCCCAUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGCAA-UGGAGCGUCGAAUUCUGGGUGCCACGUGGCCACUUUGGGCCCCAUCG-- ((((((((...)))).))))......(((((..((....))(((((..(((((((((((.((.(((.(..-.).))).))..)))))))))))...)).)))....))))).......-- ( -42.00, z-score = -1.96, R) >droSim1.chr2L 549925 118 + 22036055 GUACGUAGUACUUGCGGUACAAAAUAGCCCAUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGCAAGCUGAGCGUCGAAUUCUGGGUGCCACGUGGCCACUUUGGGCCCCAUCG-- ((((((((...)))).))))......(((((..((....))(((((..(((((((((((.((.(((........))).))..)))))))))))...)).)))....))))).......-- ( -41.80, z-score = -1.87, R) >consensus GUACGUAGUACUUGCGAUACAAAAUAGCCCGUAGAAUAAUCGGCACCUGCACCCAGAAUGGAAGCUGAAA__GGAGCGUCGAAUUCUGGGUGCCACGUGGCCACUUUGGGCCUCAUCG_A ((((((((...))))).)))......(((((..((....))(((((..(((((((((((.((.(((........))).)).).))))))))))...)).)))....)))))......... (-28.64 = -30.00 + 1.36)

| Location | 541,428 – 541,546 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.22608 |
| G+C content | 0.51278 |
| Mean single sequence MFE | -40.61 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 541428 118 - 23011544 UUCGAUGAGGCCCAAAGUGGCCACGUGGCACCCAGAAUUCGACGCUCU--UUUCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUACGGGCUAUUUUGUAUCGCGAGUACUACGUAC ((((.(((....((((((((((.(((((((((((((((..((.(((..--....))).)).))))))))))).......((....))))))))))))))))..))))))).......... ( -44.80, z-score = -3.45, R) >droEre2.scaffold_4929 596475 118 - 26641161 UUUGAAGAGCCCCAAAGUGGCUACGUGGCACCCAGUAACCGAAGCUCAU-UCUCAUCUUCGAUUCUGGGUGCAUAU-CUGAUUAUUCUACGGUCUAUUUUGUAUCGCAAGUACUACGUAC .......((((.......))))(((((((((((((.((.(((((.....-......))))).))))))))))....-.......................(((.(....))))))))).. ( -31.30, z-score = -1.61, R) >droYak2.chr2L 528730 118 - 22324452 UAUGAUGAGCCCCAAAGUGGCUACGUGGCACCCAGAAGCCGACGCUCCC-UUCCAUCUUCGAUUCUGGGUGCAUGU-CCGAUUGUUCUACGGGCUAUUUUGUAUCGCAAGUACUACGUAC .......((((.......))))(((((((((((((((..(((.......-........))).))))))))))..((-(((.........)))))......(((.(....))))))))).. ( -36.36, z-score = -1.51, R) >droSec1.super_14 524000 117 - 2068291 --CGAUGGGGCCCAAAGUGGCCACGUGGCACCCAGAAUUCGACGCUCCA-UUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUAUGGGCUAUUUUGUACCGCAAGUACUACGUAC --...((.((..((((((((((.(((((((((((((((..((.(((...-....))).)).))))))))))).......((....))))))))))))))))..)).)).((((...)))) ( -45.00, z-score = -2.54, R) >droSim1.chr2L 549925 118 - 22036055 --CGAUGGGGCCCAAAGUGGCCACGUGGCACCCAGAAUUCGACGCUCAGCUUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUAUGGGCUAUUUUGUACCGCAAGUACUACGUAC --...((.((..((((((((((.(((((((((((((((..((.((((.....).))).)).))))))))))).......((....))))))))))))))))..)).)).((((...)))) ( -45.60, z-score = -2.44, R) >consensus U_CGAUGAGGCCCAAAGUGGCCACGUGGCACCCAGAAUUCGACGCUCA__UUGCAGCUUCCAUUCUGGGUGCAGGUGCCGAUUAUUCUACGGGCUAUUUUGUAUCGCAAGUACUACGUAC ......((....((((((((((.((((((((((((((...((.(((........))).))..)))))))))).......((....))))))))))))))))..))....((((...)))) (-34.62 = -34.46 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:08 2011