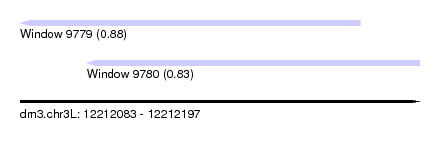
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,212,083 – 12,212,197 |
| Length | 114 |
| Max. P | 0.884377 |

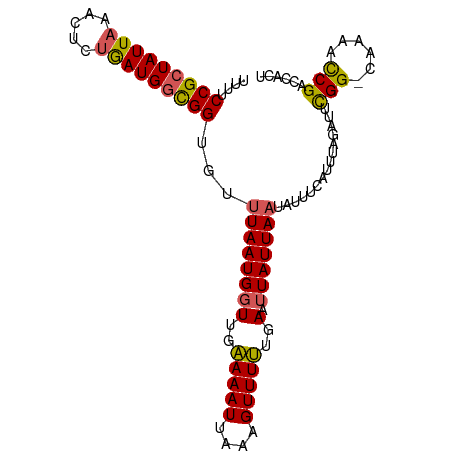
| Location | 12,212,083 – 12,212,180 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Shannon entropy | 0.21833 |
| G+C content | 0.30457 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -15.18 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12212083 97 - 24543557 UUUUCCGCUAUUAAACUCUAAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUACUAUUUCAUUUAGAUUCGG-CCAAUCCGACCACU ....(((((((((.....)))))))))....((((((((.((((.......)))).))))))))(((.......)))..((((-.....))))..... ( -20.10, z-score = -1.91, R) >droYak2.chr3L 12262736 97 - 24197627 UUUUCCGCUAUUAAACUCCGAUGGUGGUUUUUAAUGGUCGUAAAUUAAAGUUUGUAAAUUAUUAAUAUUUCAUUUAGUUACGG-CACACUCGACCAGU ....((((((((.......)))))))).......((((((........(((.(((.....(((((........)))))....)-)).))))))))).. ( -20.00, z-score = -1.76, R) >droEre2.scaffold_4784 12201878 97 - 25762168 UUUUCCGCUAUUAAACUCUAAUGUUGGCGUUUAAUGGUUGUAAAUUAAAGUUUGUGAAAUAUUAAUAUUUCAUUUAGAUAUGG-AAAAACCGACCUCU ((((((((((((((((.(((....))).))))))))))...........((((((((((((....))))))))..))))..))-)))).......... ( -21.20, z-score = -2.98, R) >droSec1.super_0 4397131 98 - 21120651 UUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGAUUCGGUUUAAACCGACCAUU ....(((((((((.....)))))))))...(((((((((.((((.......)))).)))))))))..............(((((....)))))..... ( -23.00, z-score = -2.75, R) >droSim1.chr3L 11586131 97 - 22553184 UUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGAUUCGG-GCAAACCGACCUCU ....(((((((((.....)))))))))...(((((((((.((((.......)))).)))))))))..........(((.((((-.....))))..))) ( -23.20, z-score = -2.48, R) >consensus UUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGAUUCGG_CAAAACCGACCACU ....(((((((((.....)))))))))...((((((((..(((((....)))))..).)))))))...............(((......)))...... (-15.18 = -14.70 + -0.48)

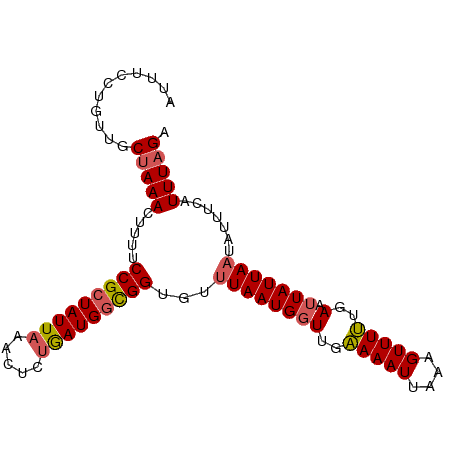
| Location | 12,212,102 – 12,212,197 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Shannon entropy | 0.14229 |
| G+C content | 0.27579 |
| Mean single sequence MFE | -18.76 |
| Consensus MFE | -14.82 |
| Energy contribution | -14.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12212102 95 - 24543557 AUUUCCUGUUGCUAAAUUUUUCCGCUAUUAAACUCUAAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUACUAUUUCAUUUAGA ...........((((((....(((((((((.....)))))))))....((((((((.((((.......)))).)))))))).......)))))). ( -19.60, z-score = -2.80, R) >droYak2.chr3L 12262755 95 - 24197627 AUUUCCGGUUGCUAAACUUUUCCGCUAUUAAACUCCGAUGGUGGUUUUUAAUGGUCGUAAAUUAAAGUUUGUAAAUUAUUAAUAUUUCAUUUAGU ........((((.(((((((..((((((((((..(((....)))..)))))))).))......)))))))))))..................... ( -15.60, z-score = -0.96, R) >droEre2.scaffold_4784 12201897 95 - 25762168 AUUUCCGGUUGCAAAACUUUUCCGCUAUUAAACUCUAAUGUUGGCGUUUAAUGGUUGUAAAUUAAAGUUUGUGAAAUAUUAAUAUUUCAUUUAGA .............(((((((.(.((((((((((.(((....))).)))))))))).)......)))))))((((((((....))))))))..... ( -18.60, z-score = -2.04, R) >droSec1.super_0 4397151 95 - 21120651 AUUUCCUGUUGCUAAACUUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGA ...........(((((.....(((((((((.....)))))))))...(((((((((.((((.......)))).))))))))).......))))). ( -20.00, z-score = -2.74, R) >droSim1.chr3L 11586150 95 - 22553184 AUUUCCUGUCGCUAAACUUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGA ...........(((((.....(((((((((.....)))))))))...(((((((((.((((.......)))).))))))))).......))))). ( -20.00, z-score = -2.62, R) >consensus AUUUCCUGUUGCUAAACUUUUCCGCUAUUAAACUCUGAUGGCGGUGUUUAAUGGUUGAAAAUUAAAGUUUUUGAAUUAUUAAUAUUUCAUUUAGA ...........(((((.....(((((((((.....)))))))))...((((((((..(((((....)))))..).))))))).......))))). (-14.82 = -14.86 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:32 2011