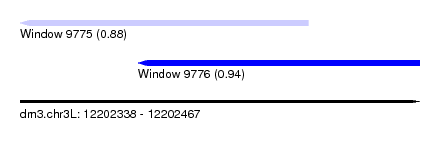
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,202,338 – 12,202,467 |
| Length | 129 |
| Max. P | 0.942261 |

| Location | 12,202,338 – 12,202,431 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Shannon entropy | 0.30369 |
| G+C content | 0.47185 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -18.30 |
| Energy contribution | -17.86 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
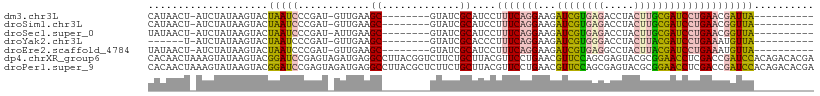
>dm3.chr3L 12202338 93 - 24543557 CGUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGAUUAUUGGAACCAUAUAUGCAUAUGUAGCCCCCGGCGAAUCA ....(((((((..(((((((...(((..((.....))..)))))))))).)))....((...(((((....)))))...))....)))).... ( -25.10, z-score = -1.68, R) >droSim1.chr3L 11573769 93 - 22553184 CGUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUAUUGGAACCAUAUAUGCAAAUGUAGCCCCCGCCGCAUCA ......((((...(((((((...(((..((.....))..)))))))))).((((.....))))....))))..(((..((....))..))).. ( -26.40, z-score = -2.07, R) >droSec1.super_0 4387522 93 - 21120651 CGUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUAUUGGAACCAUAUAUGCAUAUGUAGCCCCCGCCGCGUCA .....(((.....(((((((...(((..((.....))..)))))))))).(((....((...(((((....)))))...))...))))))... ( -26.50, z-score = -1.66, R) >droYak2.chr3L 12252852 84 - 24197627 CGUAUCGCACCCUUUCAGGAAGAUCGUGGGACCUACUUACGAUCCUGAAAUGUUAUUGGAACCAUAAAUGUGAACCCCGAAUCA--------- ....(((((...((((((((...(((((((.....)))))))))))))))......((....))....)))))...........--------- ( -20.60, z-score = -1.37, R) >droEre2.scaffold_4784 12192082 83 - 25762168 CGUAUCGCAUCCUUUCAGGAAGAUCGUGAGGCCUACUUACGAUCCUGAAAUGUUAUUGGAACCAUAAAUGU-AACCCCGAAUCA--------- ....(((.....((((((((...(((((((.....))))))))))))))).(((((((....)).....))-)))..)))....--------- ( -20.80, z-score = -2.06, R) >consensus CGUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUAUUGGAACCAUAUAUGCAAAUGUAGCCCCCG_CG__UCA ......((((...(((((((...(((((((.....)))))))))))))).......((....))...))))...................... (-18.30 = -17.86 + -0.44)
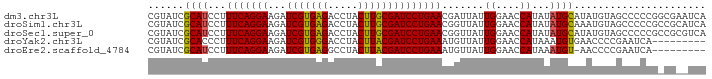
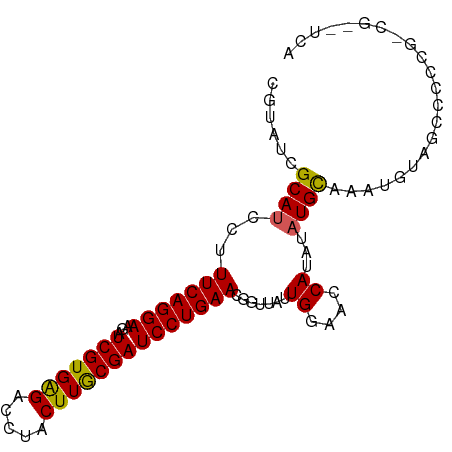
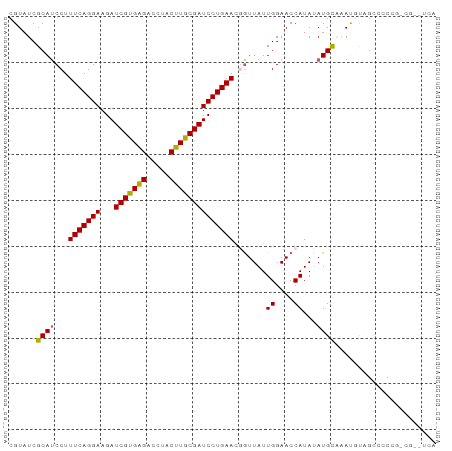
| Location | 12,202,376 – 12,202,467 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.10 |
| Shannon entropy | 0.55258 |
| G+C content | 0.44270 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -13.09 |
| Energy contribution | -12.59 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
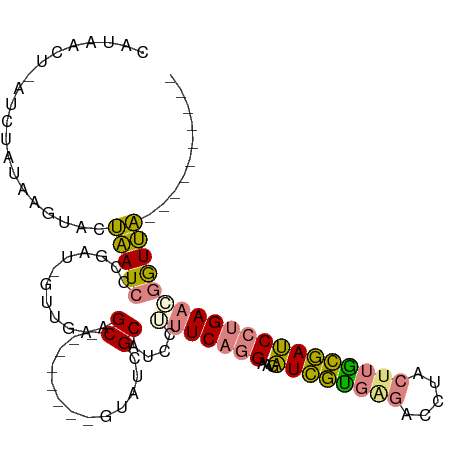
>dm3.chr3L 12202376 91 - 24543557 CAUAACU-AUCUAUAAGUACUAAUCCCGAU-GUUGAAGC--------GUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGAUUA---------- .......-............(((((..(((-((.((...--------...)))))))..(((((((...(((..((.....))..)))))))))).)))))---------- ( -21.40, z-score = -1.94, R) >droSim1.chr3L 11573807 91 - 22553184 CAUAACU-AUCUAUAAGUACUAAUCCCGAU-GUUGAAGC--------GUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUA---------- ..(((((-...................(((-((.((...--------...)))))))..(((((((...(((..((.....))..)))))))))).)))))---------- ( -21.80, z-score = -1.53, R) >droSec1.super_0 4387560 91 - 21120651 UAUAACU-AUCUAUAAGUACUAAUCCCGAU-AUUGAAGC--------GUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUA---------- .......-..................((((-((......--------))))))...((.(((((((...(((..((.....))..)))))))))).))...---------- ( -22.20, z-score = -1.95, R) >droYak2.chr3L 12252881 85 - 24197627 ------U-AUCUAUAAGUACUAAUCCCGAU-GUUGAAGC--------GUAUCGCACCCUUUCAGGAAGAUCGUGGGACCUACUUACGAUCCUGAAAUGUUA---------- ------.-..................((((-(.......--------.))))).....((((((((...(((((((.....))))))))))))))).....---------- ( -18.60, z-score = -0.83, R) >droEre2.scaffold_4784 12192110 91 - 25762168 UAUAACU-AUCUAUAAGUACUAAUCCCGAU-GUUGAAGC--------GUAUCGCAUCCUUUCAGGAAGAUCGUGAGGCCUACUUACGAUCCUGAAAUGUUA---------- ..((((.-...................(((-((.((...--------...))))))).((((((((...(((((((.....))))))))))))))).))))---------- ( -23.20, z-score = -2.46, R) >dp4.chrXR_group6 2543157 111 - 13314419 CACAACUAAAGUAUAAGUACGGAUCCGAGUAGAUGAGGCCUUACGGUCUUCUGCUUACGUUCCUGAACGUUCCAGCGAGUACGCGGAACCUCGACCGAUCCACAGACACGA ..........(((....)))(((((.(((((((..(((((....))))))))))))..(((....)))(((((.(((....)))))))).......))))).......... ( -31.50, z-score = -1.12, R) >droPer1.super_9 835384 111 - 3637205 CACAACUAAAGUAUAAGUACGGAUCCGAGUAGAUGAGGCCUUACGCUCUUCUGCUUACGUUCCUGAACGUUCCAGCGAGUACGCGGAACCUCGACCGAUCCACAGACACGA ..........(((....)))(((((.(((((((.(((........))).)))))))..(((....)))(((((.(((....)))))))).......))))).......... ( -29.30, z-score = -0.88, R) >consensus CAUAACU_AUCUAUAAGUACUAAUCCCGAU_GUUGAAGC________GUAUCGCAUCCUUUCAGGAAGAUCGUGAGACCUACUUGCGAUCCUGAACGGUUA__________ ....................(((((............((.............))....(((((((...((((((((.....)))))))))))))))))))).......... (-13.09 = -12.59 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:28 2011