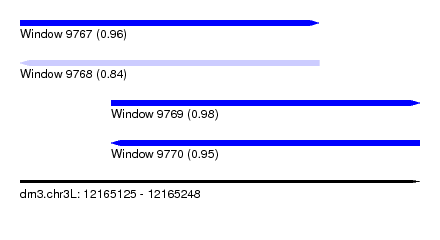
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,165,125 – 12,165,248 |
| Length | 123 |
| Max. P | 0.976603 |

| Location | 12,165,125 – 12,165,217 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Shannon entropy | 0.17647 |
| G+C content | 0.47247 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
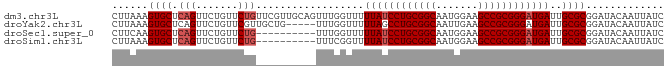
>dm3.chr3L 12165125 92 + 24543557 CUUAAAGUGCUCAGUUCUGUUCUGUUCGUUGCAGUUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUC .............(((((((.((((.....))))....((..((((((((((((.......))))))))))))..))))))).))....... ( -31.60, z-score = -2.84, R) >droYak2.chr3L 12213088 87 + 24197627 CUUAAAGUGCUCAGUUCUGUUCGUUGCUG-----UUUGGUUUUUAGCCUGCGGCAAUUGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUC ......((((...((((((((((((((((-----(..(((.....))).))))))).)))....)))))))....))))............. ( -25.80, z-score = -1.06, R) >droSec1.super_0 4350206 82 + 21120651 CUUCAAGUGCUCAGUUCUGUUCUG----------UUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUC .................(((((((----------(...((..((((((((((((.......))))))))))))..))))))).)))...... ( -28.90, z-score = -2.96, R) >droSim1.chr3L 11536445 82 + 22553184 CUUAAAGUGCUCAGUUCUGUUCUG----------UUUCGGUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUC ......((((.(((.......)))----------........((((((((((((.......))))))))))))..))))............. ( -27.30, z-score = -2.45, R) >consensus CUUAAAGUGCUCAGUUCUGUUCUG__________UUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUC ......((((.((....)).......................((((((((((((.......))))))))))))..))))............. (-24.08 = -24.32 + 0.25)
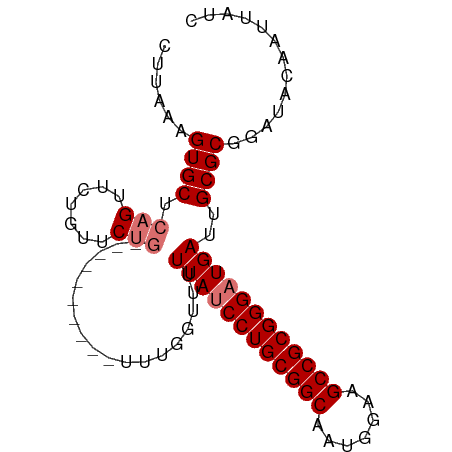
| Location | 12,165,125 – 12,165,217 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.91 |
| Shannon entropy | 0.17647 |
| G+C content | 0.47247 |
| Mean single sequence MFE | -18.85 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12165125 92 - 24543557 GAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAACUGCAACGAACAGAACAGAACUGAGCACUUUAAG ((((....)))).((.((.((((((.(((((.......))))).))))..........(((.......)))....))..)).))........ ( -20.40, z-score = -1.99, R) >droYak2.chr3L 12213088 87 - 24197627 GAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCAAUUGCCGCAGGCUAAAAACCAAA-----CAGCAACGAACAGAACUGAGCACUUUAAG ((((....)))).((((.........(((((.......))))).((.......))...-----..))......((....)).))........ ( -16.00, z-score = -0.32, R) >droSec1.super_0 4350206 82 - 21120651 GAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAA----------CAGAACAGAACUGAGCACUUGAAG ((((....))))...((....((((.(((((.......))))).))))..........----------(((.......))).))........ ( -19.50, z-score = -2.20, R) >droSim1.chr3L 11536445 82 - 22553184 GAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAACCGAAA----------CAGAACAGAACUGAGCACUUUAAG ((((....))))...((....((((.(((((.......))))).))))..........----------(((.......))).))........ ( -19.50, z-score = -2.18, R) >consensus GAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAA__________CAGAACAGAACUGAGCACUUUAAG ((((....))))...((....((((.(((((.......))))).)))).........................((....)).))........ (-16.67 = -16.93 + 0.25)
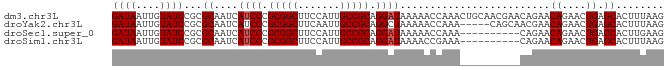
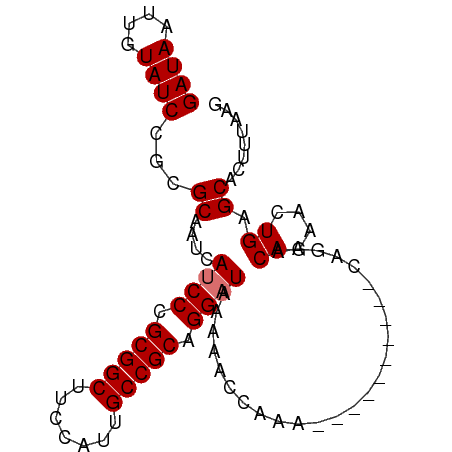
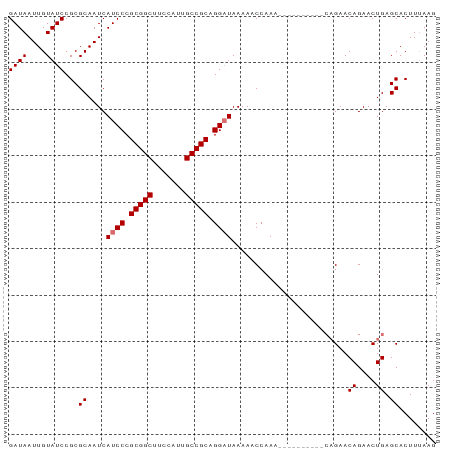
| Location | 12,165,153 – 12,165,248 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Shannon entropy | 0.14200 |
| G+C content | 0.52197 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -28.51 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12165153 95 + 24543557 UUGCAGUUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUCGGGUGAAAUGGGCGGGAAAACG .....((((.....((((((((((((.......))))))))))))((.((((((((....))))).(((.....))).......))).)))))). ( -35.40, z-score = -2.30, R) >droYak2.chr3L 12213111 93 + 24197627 UUGCUGUUUGGUUUUUAGCCUGCGGCAAUUGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGG-UGAAAUGG-CGGGAAAACG (((.((((((((..(((.((((((((.......)))))))).)))..)).)))))))))...(((.((((.((..-..)).)))-).)))..... ( -33.40, z-score = -2.16, R) >droSec1.super_0 4350229 89 + 21120651 -----GUUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGGCACUUGGCUGAAAUGG-CGGGAAAGCG -----.....((..((((((((((((.......))))))))))))..))(((((((....))))).....((((.(......).-))))...)). ( -35.80, z-score = -3.20, R) >droSim1.chr3L 11536468 89 + 22553184 -----GUUUCGGUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGGCUGAAAUGG-CGGGAAAACG -----((((.....((((((((((((.......))))))))))))((.((((((((....)))))((((....))))......)-)).)))))). ( -38.00, z-score = -3.89, R) >consensus _____GUUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGGCUGAAAUGG_CGGGAAAACG .....((((.....((((((((((((.......))))))))))))((.((((((((....)))))((((....))))......).)).)))))). (-28.51 = -29.32 + 0.81)

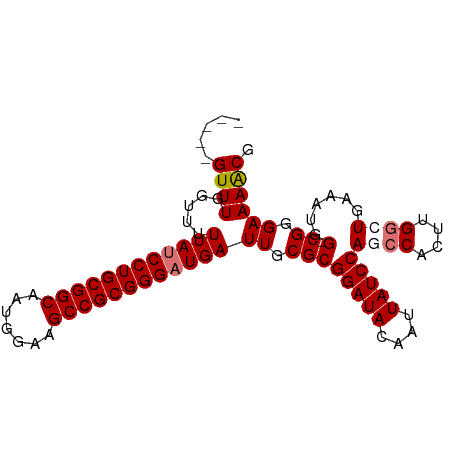
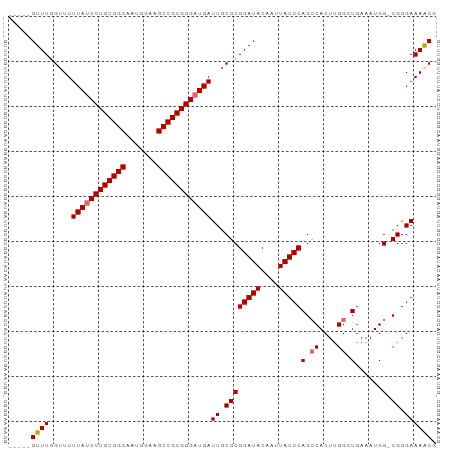
| Location | 12,165,153 – 12,165,248 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Shannon entropy | 0.14200 |
| G+C content | 0.52197 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12165153 95 - 24543557 CGUUUUCCCGCCCAUUUCACCCGAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAACUGCAA .(((((..(((((((((.....))))))..(((((....)))))))).....((((.(((((.......))))).)))).))))).......... ( -28.20, z-score = -2.17, R) >droYak2.chr3L 12213111 93 - 24197627 CGUUUUCCCG-CCAUUUCA-CCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCAAUUGCCGCAGGCUAAAAACCAAACAGCAA .(((((.(((-((((((..-..))))))).((((.(((((......))))).)))).(((((.......))))).))...))))).......... ( -28.50, z-score = -2.74, R) >droSec1.super_0 4350229 89 - 21120651 CGCUUUCCCG-CCAUUUCAGCCAAGUGCCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAAC----- .((......)-)............((((..(((((....)))))))))....((((.(((((.......))))).))))...........----- ( -25.20, z-score = -2.30, R) >droSim1.chr3L 11536468 89 - 22553184 CGUUUUCCCG-CCAUUUCAGCCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAACCGAAAC----- .(((((..((-(......((((....))))(((((....)))))))).....((((.(((((.......))))).)))))))))......----- ( -29.70, z-score = -2.98, R) >consensus CGUUUUCCCG_CCAUUUCAGCCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAAC_____ .......(((.((((((.....)))))).)))...(((((......))))).((((.(((((.......))))).))))................ (-22.64 = -22.95 + 0.31)

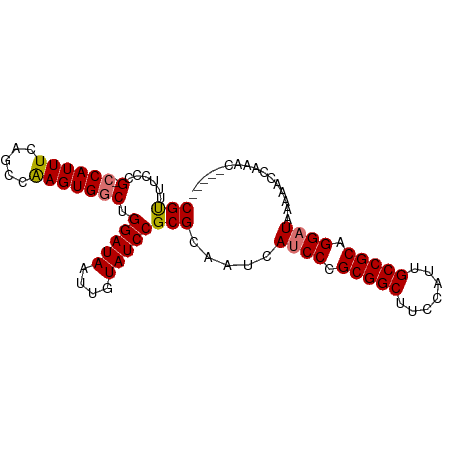
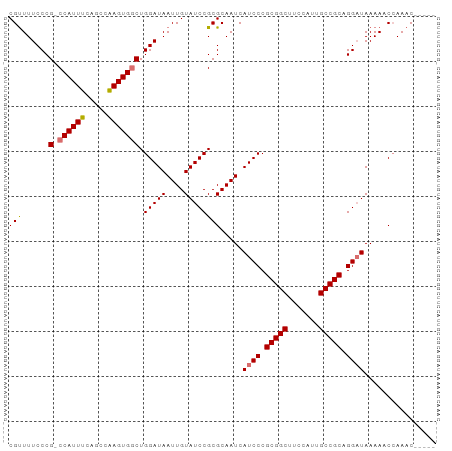
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:23 2011