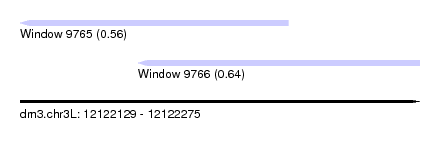
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,122,129 – 12,122,275 |
| Length | 146 |
| Max. P | 0.641880 |

| Location | 12,122,129 – 12,122,227 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 57.32 |
| Shannon entropy | 0.74947 |
| G+C content | 0.43560 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -8.44 |
| Energy contribution | -8.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12122129 98 - 24543557 ---GGUGAAGCAGCUGGAAGAUCCAGUUAAGAUUU---CCUCGCUCCAGAGACACCUUCUCCAGCUGUCUAAAUA-GGAGAGUC----UAAAAAAAAGAAUGCCCAAAA ---((((....((((((.....)))))).......---.(((......))).))))((((((.............-))))))..----..................... ( -21.82, z-score = -0.48, R) >droAna3.scaffold_13337 3828641 108 + 23293914 UUUCCAUAAGUUUUCAAAAUAAUUGAUUAAUCGUUAAAUACUAUAAGGAAAAAGGUGAUAUUCUCUACGUUAGUU-CACAUUUCUUUCCAUUGCAAUGAGAGCUGUGUA ....((((.(((((((......(((((.....))))).........(((((...((((................)-))).....))))).......))))))))))).. ( -16.19, z-score = -0.16, R) >droYak2.chr3L 12170164 106 - 24197627 ---GCUGCAGCAGCUGGAGGAUCCACAUACGCCUCCAGCUGCGCUCCAUAGGCACCCCCUCCACCUGUCCAAAUAUCACGAGUUGCAGUCUAUAAAAAAAUGCCAAAAA ---(((((((((((((((((...........)))))))))))((((.(((((...........)))))...........)))))))))).................... ( -33.40, z-score = -3.89, R) >droSec1.super_0 4307197 98 - 21120651 ---GCUGCAGCAGCUGGAAGAUCCAGUUCAUAUUU---CCUCGCUCCAGUGACACCUUCUCCAGCUGUCCAAAUA-GGUGAGUC----UAUAAAAAGGAAUGCCCAAAA ---......(((((((((.((......))......---..((((....)))).......))))))))).......-((((..((----(......)))..))))..... ( -21.90, z-score = -0.37, R) >droSim1.chr3L 11493344 97 - 22553184 ---GCUGCAGCAGCUGGAAGAUCCAGUUCAGAUUU---CCUCGCUCCAGAGACA-CUUCUACAGCUGUCCAAAUA-GGUGAGUC----UAUAAAAAGGAAUGCCCAAAA ---...(((((((((((.....)))))).(((...---.(((......)))...-..)))...))))).......-((((..((----(......)))..))))..... ( -21.30, z-score = 0.25, R) >consensus ___GCUGCAGCAGCUGGAAGAUCCAGUUAAGAUUU___CCUCGCUCCAGAGACACCUUCUCCAGCUGUCCAAAUA_GGCGAGUC____UAUAAAAAAGAAUGCCCAAAA .........((((((((.....)))))............((((((.....((((.((.....)).)))).......))))))..................)))...... ( -8.44 = -8.36 + -0.08)

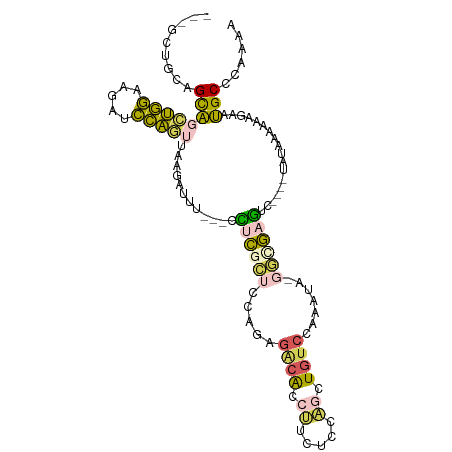
| Location | 12,122,172 – 12,122,275 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Shannon entropy | 0.32385 |
| G+C content | 0.47925 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12122172 103 - 24543557 UACAUCAUUAUCUAAAUUCAAGAGUGAGCCACUAGAAACUGGUGGUUAGGUGAAGCAGCUGGAAGAUCCAGUUAAGAUUU---CCUCGCUCCAGAGACACCUUCUC .....................(((..(((((((((...)))))))))(((((....((((((.....)))))).......---.(((......))).))))).))) ( -28.40, z-score = -1.48, R) >droYak2.chr3L 12170212 103 - 24197627 ---GGCAUUAUCUAAACGCAAGAGUACGCCACUGGGAACUGGCUAUUAGCUGCAGCAGCUGGAGGAUCCACAUACGCCUCCAGCUGCGCUCCAUAGGCACCCCCUC ---(((....(((.......)))....)))...(((...((.((((.....((.(((((((((((...........)))))))))))))...)))).))..))).. ( -38.80, z-score = -2.46, R) >droSec1.super_0 4307240 103 - 21120651 CACAUCAUUAUCUAAAUUCAAGAGUGAGCCACUAUAAACUGGUGGCUAGCUGCAGCAGCUGGAAGAUCCAGUUCAUAUUU---CCUCGCUCCAGUGACACCUUCUC .................(((.(((((((....(((.((((((...(((((((...))))))).....)))))).)))...---.)))))))...)))......... ( -29.20, z-score = -2.25, R) >droSim1.chr3L 11493387 102 - 22553184 CACAUCAUUAUCUAAAUUCAAGAGUGAGCCACUAGAAACUGGUGGUUAGCUGCAGCAGCUGGAAGAUCCAGUUCAGAUUU---CCUCGCUCCAGAGACACUUCUA- .....................((((((((((((((...)))))))))..(((.(((((((((.....))))))..((...---..))))).)))...)))))...- ( -28.70, z-score = -1.44, R) >consensus CACAUCAUUAUCUAAAUUCAAGAGUGAGCCACUAGAAACUGGUGGUUAGCUGCAGCAGCUGGAAGAUCCAGUUAAGAUUU___CCUCGCUCCAGAGACACCUCCUC .....................(((((((.....(((((((((...(((((((...))))))).....))))))....)))....)))))))............... (-18.67 = -18.43 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:21 2011