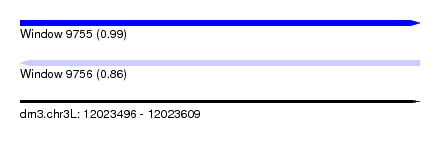
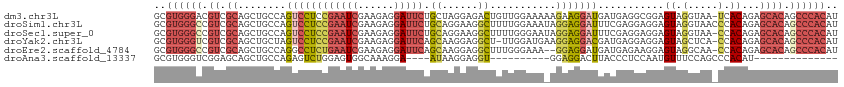
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,023,496 – 12,023,609 |
| Length | 113 |
| Max. P | 0.987952 |

| Location | 12,023,496 – 12,023,609 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.52432 |
| G+C content | 0.56423 |
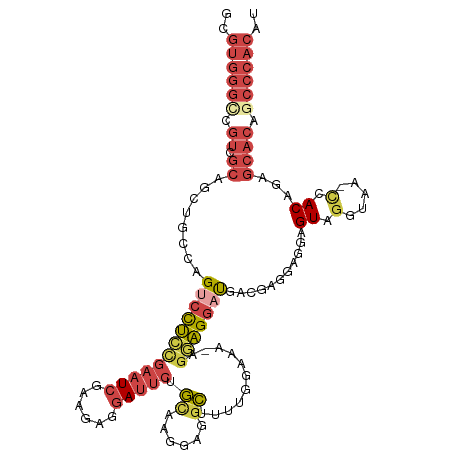
| Mean single sequence MFE | -37.59 |
| Consensus MFE | -16.39 |
| Energy contribution | -18.57 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
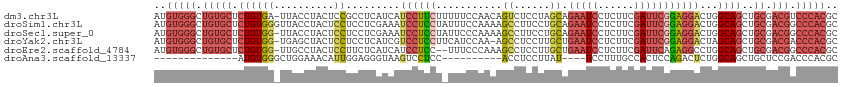
>dm3.chr3L 12023496 113 + 24543557 AUGUGGGCUGUGCUCUGUGA-UUACCUACUCCGCCUCAUCAUCCUUCUUUUUCCAACAGUCUCCUAGCAGAAUCCUCUUCGAUUCGGAGGACUGGCAGCUGCGACGUCCCACGC ..(((((.(((((.((((((-(...............)))................((((((((.....(((((......)))))))).)))))))))..)).))).))))).. ( -29.26, z-score = -0.27, R) >droSim1.chr3L 11393223 114 + 22553184 AUGUGGGCUGUGCUCUGUGGGUUACCUACUCCUCCUCGAAAUCCUCCUAUUUCCAAAAGCCUUCCUGCAGAAUCCUCUUCGAUUCGGAGGACUGGCAGCUGCGACGGCCCACGC ..(((((((((((.((((((((.....))))..........((((((...........((......)).(((((......)))))))))))...))))..)).))))))))).. ( -42.00, z-score = -3.13, R) >droSec1.super_0 4209474 113 + 21120651 AUGUGGGCUGUGCUCUGUGG-UUACCUACUCCUCCUCGAAAUCCUCCUAUUCCCAAAAGCCUUCCUGCAGAAUCCUCUUCGAUUCGGAGGACUGGCAGCUGCGACGGCCCACGC ..(((((((((....((..(-((.((...((((((.......................((......)).(((((......)))))))))))..)).)))..))))))))))).. ( -41.20, z-score = -3.46, R) >droYak2.chr3L 12069738 112 + 24197627 AUGUGGGCUGUGCUCUGUGG-UGAGCUACUCCUCCUCAUCGUCCUCCUUCAUCCAA-AGCCUCCUUGCUGAAUCCUCUUCGAUUCGGAGGACUAGCAGCUGCGACGACCCACGC ..(((((.(((((.((((((-((((.........))))))(((((((.........-(((......)))(((((......))))))))))))..))))..)).))).))))).. ( -42.90, z-score = -3.71, R) >droEre2.scaffold_4784 12011313 111 + 25762168 AUGUGGGCUGUGCUCUGUGG-UUGCCUACUCCUUCUCAUCAUCCUCC--UUUCCCAAAGCCUCCUUGCUGAAUCCUCUUCGAUUCAGAGGCCUGGCAGCUGCGACGGCCCACGC ..(((((((((....((..(-(((((....((((.............--........(((......)))(((((......))))).))))...))))))..))))))))))).. ( -40.20, z-score = -3.21, R) >droAna3.scaffold_13337 11401613 86 - 23293914 --------------AUGUGGGCUGGAAACAUUGGAGGGUAAGUCCUCC----------ACCUCCUUAU----UCCUUUGCCACUCCAGACUCUGGCAGCUGCUCCGACCCACGC --------------..(((((.((((.....(((((((....))))))----------).........----......((..((((((...)))).))..)))))).))))).. ( -30.00, z-score = -1.86, R) >consensus AUGUGGGCUGUGCUCUGUGG_UUACCUACUCCUCCUCAUAAUCCUCCU_UUUCCAAAAGCCUCCUUGCAGAAUCCUCUUCGAUUCGGAGGACUGGCAGCUGCGACGGCCCACGC ..(((((.(((((.((((((..........)).........((((((...........((......)).(((((......)))))))))))...))))..)).))).))))).. (-16.39 = -18.57 + 2.17)
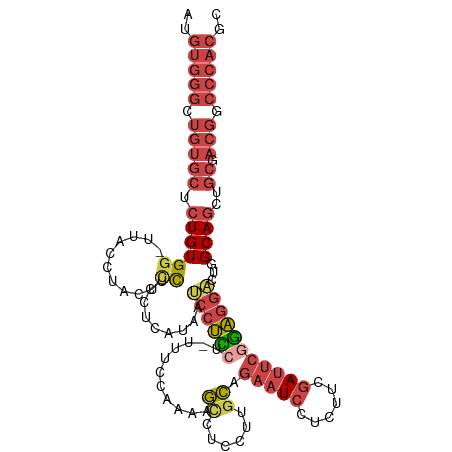
| Location | 12,023,496 – 12,023,609 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.52432 |
| G+C content | 0.56423 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -18.35 |
| Energy contribution | -20.17 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12023496 113 - 24543557 GCGUGGGACGUCGCAGCUGCCAGUCCUCCGAAUCGAAGAGGAUUCUGCUAGGAGACUGUUGGAAAAAGAAGGAUGAUGAGGCGGAGUAGGUAA-UCACAGAGCACAGCCCACAU ..(((((..((.((..(((((.(((.((((((((......)))))...(((....)))............))).)))..))))).((......-..))...))))..))))).. ( -35.40, z-score = -0.86, R) >droSim1.chr3L 11393223 114 - 22553184 GCGUGGGCCGUCGCAGCUGCCAGUCCUCCGAAUCGAAGAGGAUUCUGCAGGAAGGCUUUUGGAAAUAGGAGGAUUUCGAGGAGGAGUAGGUAACCCACAGAGCACAGCCCACAU ..((((((.((.((...(((((.(((((((((((......)))))....((((..((((((....))))))..))))..)))))).).)))).(.....).)))).)))))).. ( -41.20, z-score = -1.83, R) >droSec1.super_0 4209474 113 - 21120651 GCGUGGGCCGUCGCAGCUGCCAGUCCUCCGAAUCGAAGAGGAUUCUGCAGGAAGGCUUUUGGGAAUAGGAGGAUUUCGAGGAGGAGUAGGUAA-CCACAGAGCACAGCCCACAU ..((((((.((.((...(((((.(((((((((((......)))))....((((..((((((....))))))..))))..)))))).).)))).-(....).)))).)))))).. ( -41.20, z-score = -1.81, R) >droYak2.chr3L 12069738 112 - 24197627 GCGUGGGUCGUCGCAGCUGCUAGUCCUCCGAAUCGAAGAGGAUUCAGCAAGGAGGCU-UUGGAUGAAGGAGGACGAUGAGGAGGAGUAGCUCA-CCACAGAGCACAGCCCACAU ..((((((((((((....))..((((((((((((......)))))..(((((...))-)))......))))))))))))......((.((((.-.....))))))..))))).. ( -43.50, z-score = -2.53, R) >droEre2.scaffold_4784 12011313 111 - 25762168 GCGUGGGCCGUCGCAGCUGCCAGGCCUCUGAAUCGAAGAGGAUUCAGCAAGGAGGCUUUGGGAAA--GGAGGAUGAUGAGAAGGAGUAGGCAA-CCACAGAGCACAGCCCACAU ..((((((((((((..((.((.((((((((((((......))))).....)))))))...))...--))..).))))).......((.(....-).))........)))))).. ( -38.20, z-score = -1.26, R) >droAna3.scaffold_13337 11401613 86 + 23293914 GCGUGGGUCGGAGCAGCUGCCAGAGUCUGGAGUGGCAAAGGA----AUAAGGAGGU----------GGAGGACUUACCCUCCAAUGUUUCCAGCCCACAU-------------- ..((((((.(((((.(((.((((...))))))).))......----.........(----------(((((......)))))).....))).))))))..-------------- ( -34.90, z-score = -2.23, R) >consensus GCGUGGGCCGUCGCAGCUGCCAGUCCUCCGAAUCGAAGAGGAUUCUGCAAGGAGGCUUUUGGAAA_AGGAGGAUGACGAGGAGGAGUAGGUAA_CCACAGAGCACAGCCCACAU ..((((((.((.((....))..((((((((((((......))))).((......))...........))))))).............................)).)))))).. (-18.35 = -20.17 + 1.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:12 2011