
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,000,016 – 5,000,120 |
| Length | 104 |
| Max. P | 0.571360 |

| Location | 5,000,016 – 5,000,120 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.19 |
| Shannon entropy | 0.63609 |
| G+C content | 0.46850 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
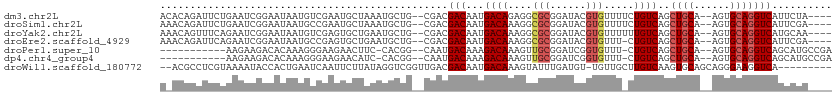
>dm3.chr2L 5000016 104 + 23011544 ACACAGAUUCUGAAUCGGAAUAAUGUCGAAUGCUAAAUGCUG--CGACGACAAUGACAGAGGCGCGGAUACGUGUUUUCUGUCAGCUGCA--AGUGCAGGUCAUUCUA---- ...(((...)))....(((((...((((...((.....))..--))))(((..(((((((((((((....)))).))))))))).((((.--...)))))))))))).---- ( -36.80, z-score = -2.80, R) >droSim1.chr2L 4855772 104 + 22036055 AAACAGAUUCUGAAUCGGAAUAAUGCCGAAUGCUAAAUGCUG--CGACGACAAUGACAAAGGCGCGGAUACGUGUUUUCUGUCAGCUGCA--AGUGCAGGUCAUUCGA---- ......((((((...)))))).....(((((((........)--)...(((..(((((((((((((....)))))))).))))).((((.--...)))))))))))).---- ( -31.50, z-score = -1.38, R) >droYak2.chr2L 11797553 104 - 22324452 AAACAGUUUCAGAAUCGGAAUAAUGUCGAGUGCUGAAUGCUG--CGACGACAAUGACAAAGGCGCGGAUACGUGUUUUUUGUCAGCUGCA--AGUGCAGGUCAUGCAA---- ...(((((((((..((((.......))))...))))).))))--....(((..(((((((((((((....)))).))))))))).((((.--...)))))))......---- ( -34.90, z-score = -1.88, R) >droEre2.scaffold_4929 5078164 103 + 26641161 AAACAGAUUCAGAAUCGGAAUAAUGCCGAGUGCUGAAUGCUG--CGACGACAAUGACAAAGGCGCGGAUACGUGUUU-CUGUCAGCUGCA--AGUGCAGGUCAUUCGA---- ...(((((((((..((((.......))))...)))))).)))--(((.(((..(((((.(((((((....)))))))-.))))).((((.--...)))))))..))).---- ( -39.50, z-score = -3.29, R) >droPer1.super_10 520280 95 + 3432795 -----------AAGAAGACACAAAGGGAAGAACUUC-CACGG--CAAUGACAAAGACAAAGUUGCGGAUCGGUGUUU-CUGUCAGCUGCA--AGUGCAGGUCAGCAUGCCGA -----------..............((((....)))-).(((--((.(((((.(((((..(........)..)))))-.))))).((((.--...)))).......))))). ( -28.00, z-score = -1.05, R) >dp4.chr4_group4 1516525 95 + 6586962 -----------AAGAAGACACAAAGGGAAGAACAUC-CACGG--CAAUGACAAAGACAAAGUUGCGGAUCGGUGUUU-CUGUCAGCUGCA--AGUGCAGGUCAGCAUGCCGA -----------..............(((......))-).(((--((.(((((.(((((..(........)..)))))-.))))).((((.--...)))).......))))). ( -26.40, z-score = -0.86, R) >droWil1.scaffold_180772 1785792 100 + 8906247 --ACGCCUCGUAAAAUACCACUGAAUCAAUUCUUAUAGGUCGGUUGACGACAAUGACAAAGUAUUUGAUGU-UGUUGCUUGUCAAGUGCAGCAGGGAAGGUCA--------- --...............((.(((...............((((.....)))).........((((((((((.-.......))))))))))..))))).......--------- ( -20.20, z-score = 0.87, R) >consensus A_ACAGAUUCUGAAUCGGAAUAAUGUCGAGUGCUAAAUGCUG__CGACGACAAUGACAAAGGCGCGGAUACGUGUUU_CUGUCAGCUGCA__AGUGCAGGUCAUUCUA____ ................................................(((...((((....(((......))).....))))..((((......))))))).......... (-11.02 = -11.57 + 0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:44 2011