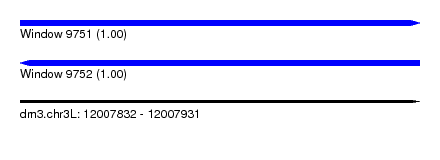
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,007,832 – 12,007,931 |
| Length | 99 |
| Max. P | 0.999390 |

| Location | 12,007,832 – 12,007,931 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.39919 |
| G+C content | 0.54440 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -28.89 |
| Energy contribution | -30.45 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
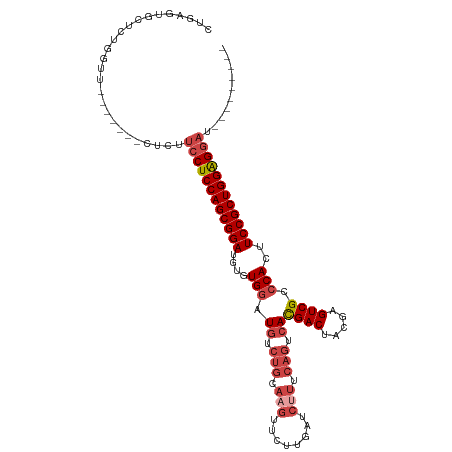
>dm3.chr3L 12007832 99 + 24543557 CUGAGUGCUCUGGUU-------CUCUUCCUCCAGCGGAUAUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGGGGAU--------- ..(((.((....)).-------))).((((((((((((...((((.((.(((.(((........))).))).))((((.....)))).)))).)))))))))))).--------- ( -43.40, z-score = -4.20, R) >droSim1.chr3L 11377662 99 + 22553184 CUGAGUGCUCUGGUU-------CUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACCACGAGUCGCCCACUUCCGCUGGAGGAU--------- ..(((.((....)).-------))).((((((((((((.(((.((.((.(((.(((........))).))).))((((.....))))))))).)))))))))))).--------- ( -43.90, z-score = -4.02, R) >droSec1.super_0 4193907 99 + 21120651 CUGAGUGCUCUGGUU-------CUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGAGGAU--------- ..(((.((....)).-------))).((((((((((((.(((.((.((.(((.(((........))).))).))((((.....))))))))).)))))))))))).--------- ( -44.60, z-score = -4.43, R) >droYak2.chr3L 12053870 106 + 24197627 CUGAGUGCUCUGGUUUCUGGUUCUCUUCCUCCAGCGGAUGUAUGCAUGUCUGGAAGUUCUUGAUCUUUCACUCACGACUAUGAGUCGCCCACUUCCGCUGGAGGAU--------- ..(((.((.(........))).))).((((((((((((.((..((.((..((((((........))))))..)).(((.....)))))..)).)))))))))))).--------- ( -40.40, z-score = -3.30, R) >droEre2.scaffold_4784 11995453 98 + 25762168 -UGAGCGCUCUGGUU-------CUCUUGCUCCAGCGGAUACAUGGCUGUCUGGAAGCUCUUGUUCCUUCAGUCAUGACUAUGAGUCGCCCACUUCCGCUGGAGGAU--------- -.(((.((....)).-------)))...((((((((((...(((((((...((((.(....)))))..)))))))(((.....))).......))))))))))...--------- ( -38.80, z-score = -2.46, R) >droAna3.scaffold_13337 11387366 90 - 23293914 ---------------------UGAACACCUCCAGCGGAUGUCUGGAUACCCCG----UUCUGGACUGGCAGUCACGACCACCAGUCGCCCACUUCCGCUGGAGGAUGUGCGACUA ---------------------.(.((((((((((((((.((..((.....)).----.....((((((..((....))..))))))....)).))))))))))).))).)..... ( -38.00, z-score = -3.19, R) >consensus CUGAGUGCUCUGGUU_______CUCUUCCUCCAGCGGAUGUGUGGAUGUCUGCAAGUUCUUGAUCUUUCAGUCACGACUACGAGUCGCCCACUUCCGCUGGAGGAU_________ ..........................((((((((((((....(((.((.(((.(((........))).))).))((((.....)))).)))..)))))))))))).......... (-28.89 = -30.45 + 1.56)

| Location | 12,007,832 – 12,007,931 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.39919 |
| G+C content | 0.54440 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -32.22 |
| Energy contribution | -33.63 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
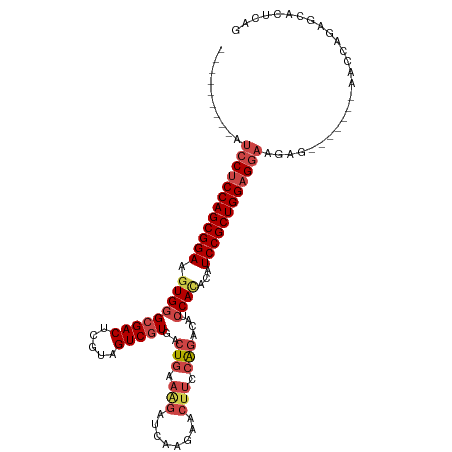
>dm3.chr3L 12007832 99 - 24543557 ---------AUCCCCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACAUAUCCGCUGGAGGAAGAG-------AACCAGAGCACUCAG ---------.(((.((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...)))))))).))).(((-------..........))).. ( -36.40, z-score = -3.41, R) >droSim1.chr3L 11377662 99 - 22553184 ---------AUCCUCCAGCGGAAGUGGGCGACUCGUGGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACACAUCCGCUGGAGGAAGAG-------AACCAGAGCACUCAG ---------.((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...)))))))))))).(((-------..........))).. ( -41.30, z-score = -3.85, R) >droSec1.super_0 4193907 99 - 21120651 ---------AUCCUCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUGCAGACAUCCACACAUCCGCUGGAGGAAGAG-------AACCAGAGCACUCAG ---------.((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...)))))))))))).(((-------..........))).. ( -41.20, z-score = -4.37, R) >droYak2.chr3L 12053870 106 - 24197627 ---------AUCCUCCAGCGGAAGUGGGCGACUCAUAGUCGUGAGUGAAAGAUCAAGAACUUCCAGACAUGCAUACAUCCGCUGGAGGAAGAGAACCAGAAACCAGAGCACUCAG ---------.((((((((((((.(((.(((((((((....))))))(.(((........))).).....))).))).)))))))))))).(((..((........).)..))).. ( -36.80, z-score = -3.35, R) >droEre2.scaffold_4784 11995453 98 - 25762168 ---------AUCCUCCAGCGGAAGUGGGCGACUCAUAGUCAUGACUGAAGGAACAAGAGCUUCCAGACAGCCAUGUAUCCGCUGGAGCAAGAG-------AACCAGAGCGCUCA- ---------...((((((((((....((((((.....))).((.(((.(((........))).))).))))).....))))))))))...(((-------..(....)..))).- ( -33.70, z-score = -1.82, R) >droAna3.scaffold_13337 11387366 90 + 23293914 UAGUCGCACAUCCUCCAGCGGAAGUGGGCGACUGGUGGUCGUGACUGCCAGUCCAGAA----CGGGGUAUCCAGACAUCCGCUGGAGGUGUUCA--------------------- .....(.(((.(((((((((((.((..(.((((((..((....))..)))))))...)----)((.....)).....)))))))))))))).).--------------------- ( -44.40, z-score = -3.63, R) >consensus _________AUCCUCCAGCGGAAGUGGGCGACUCGUAGUCGUGACUGAAAGAUCAAGAACUUCCAGACAUCCACACAUCCGCUGGAGGAAGAG_______AACCAGAGCACUCAG ..........((((((((((((.(((((((((.....)))))..(((.(((........))).)))....))))...)))))))))))).......................... (-32.22 = -33.63 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:08 2011