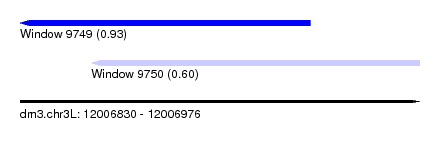
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,006,830 – 12,006,976 |
| Length | 146 |
| Max. P | 0.933166 |

| Location | 12,006,830 – 12,006,936 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.88 |
| Shannon entropy | 0.12497 |
| G+C content | 0.43696 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -28.62 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
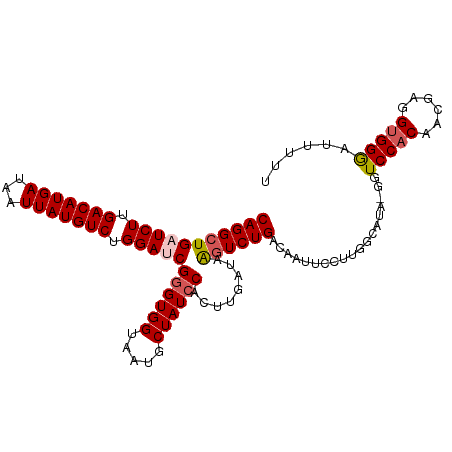
>dm3.chr3L 12006830 106 - 24543557 CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCAUA-GGUCCACAACGAGGAGGGGUUUUU (((((((((((.(((((((...))))))).)))))((((((.....))))))........))))))((..(((((((.....-.........)))))))..)).... ( -36.74, z-score = -2.91, R) >droSim1.chr3L 11376632 107 - 22553184 CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUACACUUGAUAAGUCUGACAAUUCCUUGGCAUAGGGUCCACAAAGAGGUGGGAUGUUU (((((((((((.(((((((...))))))).)))))(((((...........)))))....))))))(((..((((......))))(((((......))))).))).. ( -31.50, z-score = -1.53, R) >droSec1.super_0 4192869 107 - 21120651 CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCAUAGGGUCCACAAUGAGGUGGGAUUUUU (((((((((((.(((((((...))))))).)))))((((((.....))))))........)))))).....((((......))))(((((......)))))...... ( -35.90, z-score = -2.57, R) >droYak2.chr3L 12052748 106 - 24197627 CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGACCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUACUUUGGCUUA-GGUCCACAACGAGGUGGGAUUUUU ....((.((((((((((((...)))))))((((((((((((.....))))))......((((((............))))))-))))))....))))).))...... ( -33.90, z-score = -2.62, R) >droEre2.scaffold_4784 11994359 105 - 25762168 CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGACCGGGUGGUAAUGCUAUCCACUUGAUAGCUCUGACAAUACUUCGGCUUA-GGUCCACAACGAGGUGGAUUUUU- .((((((((((.(((((((...))))))).)))...((((.((..((((((.....))))))..))....)))))))))))(-(((((((......))))))))..- ( -32.40, z-score = -2.21, R) >consensus CAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCAUA_GGUCCACAACGAGGUGGGAUUUUU (((((((((((.(((((((...))))))).)))))((((((.....))))))........))))))...................(((((......)))))...... (-28.62 = -29.30 + 0.68)

| Location | 12,006,856 – 12,006,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Shannon entropy | 0.11330 |
| G+C content | 0.43000 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -28.58 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
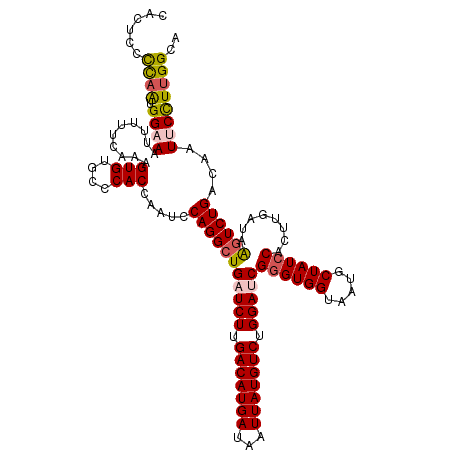
>dm3.chr3L 12006856 120 - 24543557 CACUCCCCAAUGGAAAUUUUUCAAAGUGUGCGCACCAAUCCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCA ......((((.((((..........(((....))).....(((((((((((.(((((((...))))))).)))))((((((.....))))))........))))))....)))))))).. ( -37.80, z-score = -2.63, R) >droSim1.chr3L 11376659 120 - 22553184 CACUUCCUAAUGGAAAAUUUUCAAAGUGUGCCCACCAAACCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUACACUUGAUAAGUCUGACAAUUCCUUGGCA ...((((....)))).....((((.(((((........((((..(((((((.(((((((...))))))).))))))).)))).......)))))))))...(((............))). ( -30.16, z-score = -1.05, R) >droSec1.super_0 4192896 120 - 21120651 CACUUCCUAAUGGAACUUUUUCAAAGUGUGCCCACCAAUCCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCA (((((((....)))..........))))((((........(((((((((((.(((((((...))))))).)))))((((((.....))))))........))))))..........)))) ( -34.17, z-score = -1.91, R) >droYak2.chr3L 12052774 120 - 24197627 CACUCCUCAGUGGAAAUUUUUCAAAGUGUGCCCACCAAUCCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGACCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUACUUUGGCU .(((..((((((((..........(((((..(((((..(((((((..(((.......)))......)))))))...)))))..))))).))))).)))..)))....(((....)))... ( -32.40, z-score = -1.09, R) >droEre2.scaffold_4784 11994384 120 - 25762168 CACUGCCCAAUGGAAAUUUUUCAAAGUGUGCCCACCAAUCCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGACCGGGUGGUAAUGCUAUCCACUUGAUAGCUCUGACAAUACUUCGGCU ....(((...((((.....))))((((((..(((((..(((((((..(((.......)))......)))))))...)))))....((((((.....)))))).......)))))).))). ( -34.90, z-score = -1.99, R) >consensus CACUCCCCAAUGGAAAUUUUUCAAAGUGUGCCCACCAAUCCAGGCUGAUCUUGACAUGAUAAUUAUGUCUGGAUCGGGUGGUAAUGCUAUCCACUUGAUAAGUCUGACAAUUCCUUGGCA ......((((.((((..........(((....))).....(((((((((((.(((((((...))))))).)))))((((((.....))))))........))))))....)))))))).. (-28.58 = -29.02 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:07 2011