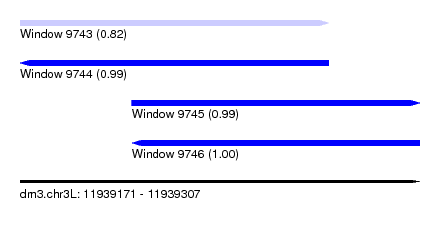
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,939,171 – 11,939,307 |
| Length | 136 |
| Max. P | 0.999522 |

| Location | 11,939,171 – 11,939,276 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.99 |
| Shannon entropy | 0.45040 |
| G+C content | 0.39403 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -17.65 |
| Energy contribution | -16.99 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11939171 105 + 24543557 --CGAUUUGACACUUCGCUGAUUGUUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUAAAU---------UGAGUGGGUUCUC--CUGAUAUCUAUGCACCAAUCGAUCUC --..............(((..(((........))).)))...((..((((.((((((((((.......---------.(((......)))--......)))).)))))).))))..)) ( -24.74, z-score = -1.00, R) >droSim1.chr3L 11307947 105 + 22553184 --UGAUUUGACACUUCGCUGAUUGUUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAU---------UGAGUGGGUUUUC--CUGAUAUCUAUGCACCAAUCGAUCUC --((((..((.((..........)).))..))))........((..((((.((((((((((.......---------....((((....)--)))...)))).)))))).))))..)) ( -23.64, z-score = -0.49, R) >droSec1.super_0 4127805 105 + 21120651 --CGAUUUGACACUUCGCUGAUUGUUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAU---------UGAGUGGGUUUUC--CUGAUAUCUAUGCACCAAUCGAUCUC --..............(((..(((........))).)))...((..((((.((((((((((.......---------....((((....)--)))...)))).)))))).))))..)) ( -23.54, z-score = -0.39, R) >droEre2.scaffold_4784 11929871 105 + 25762168 --CGAUUUGACACUCCGCUGAUUGUUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAU---------UGAGUGGGGCUUC--UUGAUAUCUAUGCACCAAUCGAUCUC --.(((..((((.((....)).))))....))).........((..((((.((((((((((.....((---------..((........)--)..)).)))).)))))).))))..)) ( -25.40, z-score = -0.69, R) >droYak2.chr3L 11984767 105 + 24197627 --CGAUUUGACACUUCGCUGAUUGUUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAU---------UGAGUGGGUUGUC--UUGAUAUCUAUGCACCAAUCGAUCUC --..............(((..(((........))).)))...((..((((.((((((((((.....((---------..((........)--)..)).)))).)))))).))))..)) ( -25.30, z-score = -0.73, R) >droAna3.scaffold_13337 6191338 96 + 23293914 -----------UAUCUGUUGAGUGGUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGAA---------AUGAUCGGAUCUCC--UUGAUAUCUAUGCACCAAUCGAUCUC -----------....((((...((((....))))..))))..((..((((.((((((((((((..(.---------..(((...)))...--)..)).)))).)))))).))))..)) ( -24.10, z-score = -1.60, R) >dp4.chrXR_group6 1929765 108 + 13314419 UGGAGAAACACUUUCUGUUGAAUGGUUCUCAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAA--------AAUUAGCGAGUCCUC--UUGAUAUCUAUGCACCAAUCGAUCUC ..(((((...(.(((....))).).)))))............((..((((.(((((((((((((((.--------..)))))(((....)--))....)))).)))))).))))..)) ( -27.50, z-score = -1.39, R) >droWil1.scaffold_180949 606536 109 - 6375548 -------CCAUCUCUUGUUGAUUGGUUCUAAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGAAUGAAAUCAAAUGAUAGAAUUCGC--AUAUUAUCUAUGCACCAAUCGAUCUC -------.........((((((((((((((.(((..((((((..........))))))....((((......)))).)))))))....((--(((.....)))))))))))))))... ( -27.40, z-score = -2.46, R) >droMoj3.scaffold_6680 9288673 108 - 24764193 --UGUUCCCAUACACCGUUGAUCGGUUCCAAUAAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGAAAU--------AAAAUGACUUUUUGGGCGUUAUCUAUGCACCAAUCGAUCUC --...........((((.....))))................((..((((.((((((((((..((...(--------((((.....)))))..))...)))).)))))).))))..)) ( -24.20, z-score = -1.32, R) >droVir3.scaffold_13049 4131602 107 - 25233164 --UGUUUUGAUGCGCCGCUGAUCGGUUCGAAUAAAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGAAAU---------GAAUUACUUUUUGGGCGAUAUCUAUGCACCAAUCGAUCUC --((((((..((.((((.....)))).)).....))))))..((..((((.((((((((((((((...(---------(((......))))..)))).)))).)))))).))))..)) ( -27.80, z-score = -1.62, R) >droGri2.scaffold_15110 22570090 107 + 24565398 --UGUGCUAAUGCGCCGCUGAUCGGUUCCAAUACAAAGCACUGAUUUCGAAUGGUGCUAGAAUUGAAAU--------UAAAUUA-UUCUUGGACGAUAUCUAUGCACCAAUCGAUCUC --.(((((..((.((((.....))))..))......))))).((..((((.((((((((((((((....--------.......-........)))).)))).)))))).))))..)) ( -26.31, z-score = -1.79, R) >consensus __CGAUUUGACACUCCGCUGAUUGGUUCUUAUCAAAAGCACUGAUUUCGAAUGGUGCUAGAGUUGAAU_________UGAGUGGGUUUUC__UUGAUAUCUAUGCACCAAUCGAUCUC ................(((.................)))...((..((((.((((((((((((((............................)))).)))).)))))).))))..)) (-17.65 = -16.99 + -0.66)

| Location | 11,939,171 – 11,939,276 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Shannon entropy | 0.45040 |
| G+C content | 0.39403 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.55 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11939171 105 - 24543557 GAGAUCGAUUGGUGCAUAGAUAUCAG--GAGAACCCACUCA---------AUUUAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAAACAAUCAGCGAAGUGUCAAAUCG-- ((..((((.((((((.((((......--(((......))).---------.......)))))))))).))))..)).(..((((((((........))))..))))..).......-- ( -26.94, z-score = -2.65, R) >droSim1.chr3L 11307947 105 - 22553184 GAGAUCGAUUGGUGCAUAGAUAUCAG--GAAAACCCACUCA---------AUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAAACAAUCAGCGAAGUGUCAAAUCA-- ((..((((.((((((.((((......--((........)).---------.......)))))))))).))))..)).(..((((((((........))))..))))..).......-- ( -24.04, z-score = -1.93, R) >droSec1.super_0 4127805 105 - 21120651 GAGAUCGAUUGGUGCAUAGAUAUCAG--GAAAACCCACUCA---------AUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAAACAAUCAGCGAAGUGUCAAAUCG-- ((..((((.((((((.((((......--((........)).---------.......)))))))))).))))..)).(..((((((((........))))..))))..).......-- ( -24.04, z-score = -1.93, R) >droEre2.scaffold_4784 11929871 105 - 25762168 GAGAUCGAUUGGUGCAUAGAUAUCAA--GAAGCCCCACUCA---------AUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAAACAAUCAGCGGAGUGUCAAAUCG-- ((..((((.((((((.((((......--(((..........---------.)))...)))))))))).))))..)).(..((((((((........)))))..)))..).......-- ( -23.10, z-score = -1.22, R) >droYak2.chr3L 11984767 105 - 24197627 GAGAUCGAUUGGUGCAUAGAUAUCAA--GACAACCCACUCA---------AUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAAACAAUCAGCGAAGUGUCAAAUCG-- ((..((((.((((((.((((......--.............---------.......)))))))))).))))..)).(..((((((((........))))..))))..).......-- ( -22.97, z-score = -1.82, R) >droAna3.scaffold_13337 6191338 96 - 23293914 GAGAUCGAUUGGUGCAUAGAUAUCAA--GGAGAUCCGAUCAU---------UUCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAACCACUCAACAGAUA----------- ((..((((.((((((.((((......--((....))((....---------.))...)))))))))).))))..))((((.((((....)))).)))).........----------- ( -26.90, z-score = -3.24, R) >dp4.chrXR_group6 1929765 108 - 13314419 GAGAUCGAUUGGUGCAUAGAUAUCAA--GAGGACUCGCUAAUU--------UUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUGAGAACCAUUCAACAGAAAGUGUUUCUCCA ((((((((.((((((.((((......--(((((........))--------)))...)))))))))).))))....((..(((((..((((.....))))...)))))..)))))).. ( -27.80, z-score = -1.75, R) >droWil1.scaffold_180949 606536 109 + 6375548 GAGAUCGAUUGGUGCAUAGAUAAUAU--GCGAAUUCUAUCAUUUGAUUUCAUUCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUUAGAACCAAUCAACAAGAGAUGG------- ((..((((.((((((.((((......--..(((.((........)).))).......)))))))))).))))..))....(((((((((......)))))).)))......------- ( -26.96, z-score = -2.06, R) >droMoj3.scaffold_6680 9288673 108 + 24764193 GAGAUCGAUUGGUGCAUAGAUAACGCCCAAAAAGUCAUUUU--------AUUUCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUUAUUGGAACCGAUCAACGGUGUAUGGGAACA-- ((..((((.((((((.((((.........((((....))))--------........)))))))))).))))..))..((.(((((((....((((.....))))..)))))))))-- ( -26.03, z-score = -1.47, R) >droVir3.scaffold_13049 4131602 107 + 25233164 GAGAUCGAUUGGUGCAUAGAUAUCGCCCAAAAAGUAAUUC---------AUUUCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUUUAUUCGAACCGAUCAGCGGCGCAUCAAAACA-- ((..((((.((((((.((((....................---------........)))))))))).))))..)).(((((......(((...))).....))))).........-- ( -25.89, z-score = -2.39, R) >droGri2.scaffold_15110 22570090 107 - 24565398 GAGAUCGAUUGGUGCAUAGAUAUCGUCCAAGAA-UAAUUUA--------AUUUCAAUUCUAGCACCAUUCGAAAUCAGUGCUUUGUAUUGGAACCGAUCAGCGGCGCAUUAGCACA-- ((..((((.((((((.((((.............-.......--------........)))))))))).))))..)).((((((((.((((....))))))).))))).........-- ( -29.69, z-score = -2.63, R) >consensus GAGAUCGAUUGGUGCAUAGAUAUCAA__GAAAACCCACUCA_________AUUCAACUCUAGCACCAUUCGAAAUCAGUGCUUUUGAUAAGAACCAAUCAGCGGAGUGUCAAAUCA__ ((..((((.((((((.((((.....................................)))))))))).))))..)).......................................... (-18.55 = -18.55 + -0.00)
| Location | 11,939,209 – 11,939,307 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.45327 |
| G+C content | 0.40257 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.39 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.994000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11939209 98 + 24543557 CUGAUUUCGAAUGGUGCUAGAGUUAAAU-UGAGUGGGUU------CUCC--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAAGUGGCAGGGCAAGUA----- ((((..((((.((((((((((.......-.(((......------))).--.....)))).)))))).))))..))))..(((((...)))))..............----- ( -28.44, z-score = -1.37, R) >droSim1.chr3L 11307985 98 + 22553184 CUGAUUUCGAAUGGUGCUAGAGUUGAAU-UGAGUGGGUU------UUCC--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAAGUGGCUGGGCAAGCU----- ((((..((((.((((((((((.......-....((((..------..))--))...)))).)))))).))))..))))..(((((...))))).((((.....))))----- ( -30.34, z-score = -1.59, R) >droSec1.super_0 4127843 98 + 21120651 CUGAUUUCGAAUGGUGCUAGAGUUGAAU-UGAGUGGGUU------UUCC--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUUGAAGUGGAUGGGCAAGUU----- .(((..((((.((((((((((.......-....((((..------..))--))...)))).)))))).))))..)))((((.((.((.....)).)).)))).....----- ( -27.24, z-score = -1.17, R) >droEre2.scaffold_4784 11929909 103 + 25762168 CUGAUUUCGAAUGGUGCUAGAGUUGAAU-UGAGUGGGGC------UUCU--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGACUCGAAGUGGGAGUGCAGCUCCUGAA ((((..((((.((((((((((.....((-..((......------..))--..)).)))).)))))).))))..))))..(((((...))))).(((((.....)))))... ( -35.90, z-score = -2.28, R) >droYak2.chr3L 11984805 98 + 24197627 CUGAUUUCGAAUGGUGCUAGAGUUGAAU-UGAGUGGGUU------GUCU--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAAGUGGUAGGGCAAGAA----- ((((..((((.((((((((((.....((-..((......------..))--..)).)))).)))))).))))..))))..(((((...)))))..............----- ( -29.00, z-score = -1.68, R) >droAna3.scaffold_13337 6191367 98 + 23293914 CUGAUUUCGAAUGGUGCUAGAAUUGAAA-UGAUCGGAUC------UCCU--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUUGAUUCGAAUUAUCCUGGCUAGCU----- ((((..((((.((((((((((((..(..-.(((...)))------...)--..)).)))).)))))).))))..)))).............................----- ( -24.90, z-score = -1.49, R) >dp4.chrXR_group6 1929805 99 + 13314419 CUGAUUUCGAAUGGUGCUAGAGUUGAAAAUUAGCGAGUC------CUCU--UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAACUGUAAUUGUAAGAA----- ((((..((((.(((((((((((((((...)))))(((..------..))--)....)))).)))))).))))..))))((((((((((........))))).)))))----- ( -29.10, z-score = -3.07, R) >droWil1.scaffold_180949 606569 108 - 6375548 CUGAUUUCGAAUGGUGCUAGAAUUGAAUGAAAUCAAAUGAUAGAAUUCGCAUAUUAUCUAUGCACCAAUCGAUCUCAGUUCUUUAAUUCGAACAAUUUGUAUGUGGUC---- .(((..((((.(((((((((((((((((...(((....)))...))))).))....)))).)))))).))))..)))((((........))))...............---- ( -26.60, z-score = -2.09, R) >droMoj3.scaffold_6680 9288711 104 - 24764193 CUGAUUUCGAAUGGUGCUAGAAUUGAAAUAAAAUGACUU------UUUGGGCGUUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGUGUGGUGUUUGAUG-- ......((((.((((((((((..((...(((((.....)------))))..))...)))).)))))).))))..(((((((........)))))))..............-- ( -27.60, z-score = -2.56, R) >droVir3.scaffold_13049 4131640 99 - 25233164 CUGAUUUCGAAUGGUGCUAGAAUUGAAAUGAA-UUACUU------UUUGGGCGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGC-UGGUGCAUA----- ......((((.((((((((((((((...((((-......------))))..)))).)))).)))))).))))...((((((........))))))((-....))...----- ( -29.70, z-score = -3.47, R) >droGri2.scaffold_15110 22570128 99 + 24565398 CUGAUUUCGAAUGGUGCUAGAAUUGAAAUUAAAUUA-UU------CUUGGACGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCAAUUUGAACUGGC-UGCUACAUA----- ......((((.((((((((((((((...........-..------......)))).)))).)))))).))))..(((((((........))))))).-.........----- ( -25.51, z-score = -3.29, R) >consensus CUGAUUUCGAAUGGUGCUAGAGUUGAAU_UGAGUGGGUU______UUCG__UGAUAUCUAUGCACCAAUCGAUCUCAGUUCUUCGAUUCGAACUGGCAGGGCAAGUA_____ ((((..((((.((((((((((...................................)))).)))))).))))..)))).................................. (-21.39 = -21.39 + -0.00)
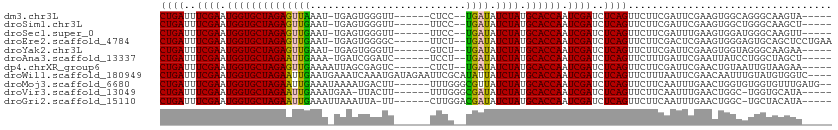
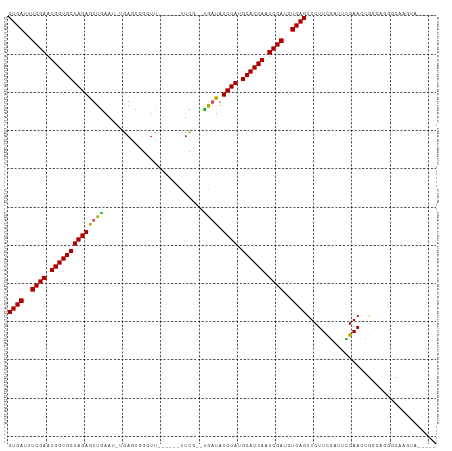
| Location | 11,939,209 – 11,939,307 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.45327 |
| G+C content | 0.40257 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.61 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11939209 98 - 24543557 -----UACUUGCCCUGCCACUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--GGAG------AACCCACUCA-AUUUAACUCUAGCACCAUUCGAAAUCAG -----..............(((((...)))))..((((..((((.((((((.((((.....--.(((------......))).-.......)))))))))).))))..)))) ( -29.24, z-score = -4.42, R) >droSim1.chr3L 11307985 98 - 22553184 -----AGCUUGCCCAGCCACUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--GGAA------AACCCACUCA-AUUCAACUCUAGCACCAUUCGAAAUCAG -----.(((.....)))..(((((...)))))..((((..((((.((((((.((((.....--.((.------.......)).-.......)))))))))).))))..)))) ( -27.34, z-score = -3.55, R) >droSec1.super_0 4127843 98 - 21120651 -----AACUUGCCCAUCCACUUCAAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--GGAA------AACCCACUCA-AUUCAACUCUAGCACCAUUCGAAAUCAG -----..............((((.....))))..((((..((((.((((((.((((.....--.((.------.......)).-.......)))))))))).))))..)))) ( -25.54, z-score = -4.15, R) >droEre2.scaffold_4784 11929909 103 - 25762168 UUCAGGAGCUGCACUCCCACUUCGAGUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--AGAA------GCCCCACUCA-AUUCAACUCUAGCACCAUUCGAAAUCAG ....((((.....))))..(((((...)))))..((((..((((.((((((.((((.....--.(((------..........-.)))...)))))))))).))))..)))) ( -30.00, z-score = -3.18, R) >droYak2.chr3L 11984805 98 - 24197627 -----UUCUUGCCCUACCACUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--AGAC------AACCCACUCA-AUUCAACUCUAGCACCAUUCGAAAUCAG -----..............(((((...)))))..((((..((((.((((((.((((.....--....------..........-.......)))))))))).))))..)))) ( -25.27, z-score = -4.27, R) >droAna3.scaffold_13337 6191367 98 - 23293914 -----AGCUAGCCAGGAUAAUUCGAAUCAAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--AGGA------GAUCCGAUCA-UUUCAAUUCUAGCACCAUUCGAAAUCAG -----...............(((........)))((((..((((.((((((.((((.....--.((.------...))((...-..))...)))))))))).))))..)))) ( -26.30, z-score = -2.63, R) >dp4.chrXR_group6 1929805 99 - 13314419 -----UUCUUACAAUUACAGUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA--AGAG------GACUCGCUAAUUUUCAACUCUAGCACCAUUCGAAAUCAG -----............((((((........))))))...((((.((((((.((((.....--.(((------((........)))))...)))))))))).))))...... ( -26.40, z-score = -3.09, R) >droWil1.scaffold_180949 606569 108 + 6375548 ----GACCACAUACAAAUUGUUCGAAUUAAAGAACUGAGAUCGAUUGGUGCAUAGAUAAUAUGCGAAUUCUAUCAUUUGAUUUCAUUCAAUUCUAGCACCAUUCGAAAUCAG ----...............((((........))))(((..((((.((((((.((((........(((.((........)).))).......)))))))))).))))..))). ( -27.36, z-score = -3.50, R) >droMoj3.scaffold_6680 9288711 104 + 24764193 --CAUCAAACACCACACCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAACGCCCAAA------AAGUCAUUUUAUUUCAAUUCUAGCACCAUUCGAAAUCAG --...............((((((........))))))...((((.((((((.((((.........((------((....))))........)))))))))).))))...... ( -24.73, z-score = -4.56, R) >droVir3.scaffold_13049 4131640 99 + 25233164 -----UAUGCACCA-GCCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCGCCCAAA------AAGUAA-UUCAUUUCAAUUCUAGCACCAUUCGAAAUCAG -----...((....-))((((((........))))))...((((.((((((.((((...........------......-...........)))))))))).))))...... ( -24.39, z-score = -3.61, R) >droGri2.scaffold_15110 22570128 99 - 24565398 -----UAUGUAGCA-GCCAGUUCAAAUUGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCGUCCAAG------AA-UAAUUUAAUUUCAAUUCUAGCACCAUUCGAAAUCAG -----.........-..((((((........))))))...((((.((((((.((((...........------..-...............)))))))))).))))...... ( -24.29, z-score = -3.22, R) >consensus _____UACUUGCCCUGCCACUUCGAAUCGAAGAACUGAGAUCGAUUGGUGCAUAGAUAUCA__AGAA______AACCCACUCA_AUUCAACUCUAGCACCAUUCGAAAUCAG ..................................((((..((((.((((((.((((...................................)))))))))).))))..)))) (-21.61 = -21.61 + 0.00)

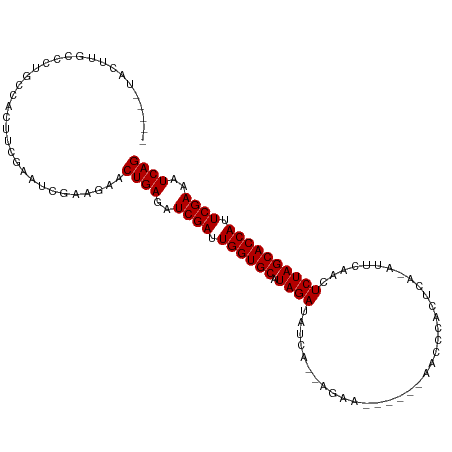
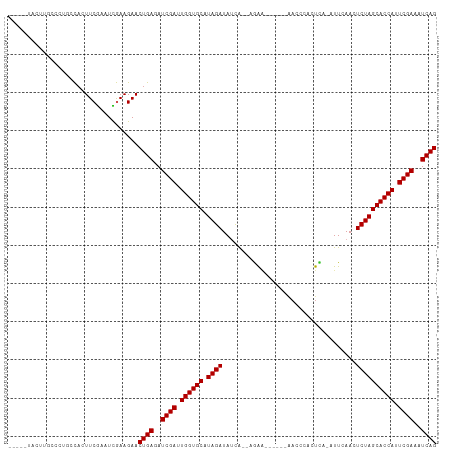
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:03 2011