
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,932,855 – 11,932,950 |
| Length | 95 |
| Max. P | 0.971078 |

| Location | 11,932,855 – 11,932,950 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.22 |
| Shannon entropy | 0.70387 |
| G+C content | 0.52233 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -11.76 |
| Energy contribution | -12.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
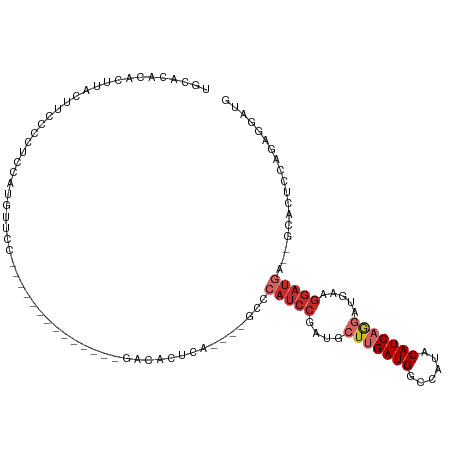
>dm3.chr3L 11932855 95 - 24543557 UGCACAUACUUACUUCCCCUCCGUGUUCC--------------GACACUCA----GCCCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA--GCACUUCAGAGUAUG ....(((((((...........((((...--------------.))))...----((.(((((....(((((((......))))))).....))))).--))......))))))) ( -30.00, z-score = -3.08, R) >droSim1.chr3L 11301565 95 - 22553184 UGCACACACUUACUUCCCCUCCGUGUUCC--------------GACACUCA----GCCCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA--GCACUCCAGAGGAUG .................((((.((((...--------------.))))...----((.(((((....(((((((......))))))).....))))).--))......))))... ( -29.10, z-score = -2.19, R) >droSec1.super_0 4121468 95 - 21120651 UGCACACACUUACUUCCCCUCUGUGUUCC--------------GACACUCA----GCCCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA--GCACUCCAGAGGAUG .................(((((((((...--------------.))))...----((.(((((....(((((((......))))))).....))))).--)).....)))))... ( -30.40, z-score = -2.39, R) >droYak2.chr3L 11978260 113 - 24197627 UGCACACACCUACUUCCCCUCCAUGUUCCGCUACCCCUGCUCCGGCACUCCCUCCGCUCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA--GCACUCCAAAGGAUG ...................(((..((.(((............))).)).......((((((((....(((((((......))))))).....))))))--))........))).. ( -30.70, z-score = -1.83, R) >droEre2.scaffold_4784 11922746 109 - 25762168 UGCACACACUUACUUCCCCUCCAUGUUCCGAUCCCCCUGCUCCGACACUCAU----CCCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA--GCACUCCAGAGGAUG .................((((..(((.(.((.(.....).)).))))(((((----(((((((((((..((.....))..)))).)))))..))))))--).......))))... ( -26.70, z-score = -0.86, R) >droAna3.scaffold_13337 6183942 93 - 23293914 --AGCAUCCUUCCAUCCCCGUCCUUUUGC--------------GACCCUUA----UCCAUUCCUAUGCAUGAUGGCCAUACAUCAAGAUGAAGGAUGA--GCACUCCGGAGGAUG --..((((((((.......((((....).--------------))).((((----(((.(((.....(.(((((......))))).)..)))))))))--)......)))))))) ( -26.60, z-score = -1.94, R) >dp4.chrXR_group6 1919093 90 - 13314419 --------CAGAAAUAUAAGCAACAUUCCACU----------CCACCUCCA-----CUCAUCCA--GGUGGAUGGCCGUACAUCAAGAUGAAGGAUGAGCGCACUCCGGAGGAUA --------........................----------...(((((.-----..(((((.--...)))))((.((.((((.........)))).)))).....)))))... ( -22.30, z-score = -0.58, R) >droPer1.super_9 214928 90 - 3637205 --------CAGAAAUAUAAGCAACAUUCCACU----------CCACCUCCA-----CUCAUCCA--GGUGGAUGGCCGUACAUCAAGAUGAAGGAUGAGCGCACUCCGGAGGAUA --------........................----------...(((((.-----..(((((.--...)))))((.((.((((.........)))).)))).....)))))... ( -22.30, z-score = -0.58, R) >droWil1.scaffold_180949 595290 77 + 6375548 ---------------------------------GAGGAAGGUUAACACCGGC---ACCAACAGCUUGCUUGAUGGCCAUACAUCAAGAUGAAGGAUGAGCAUACCCCACAGCC-- ---------------------------------..((..(((....)))..(---(((..((.....(((((((......))))))).))..)).))........))......-- ( -15.80, z-score = 0.02, R) >consensus UGCACACACUUACUUCCCCUCCAUGUUCC______________GACACUCA____GCCCAUCCGAUGCUUGAUGGCCAUACAUCAGGAUGAAGGAUGA__GCACUCCAGAGGAUG ..........................................................(((((....(((((((......))))))).....))))).................. (-11.76 = -12.80 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:00 2011