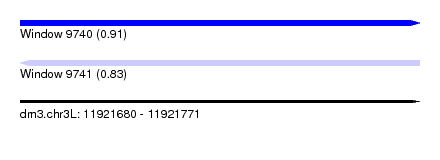
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,921,680 – 11,921,771 |
| Length | 91 |
| Max. P | 0.913862 |

| Location | 11,921,680 – 11,921,771 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Shannon entropy | 0.36114 |
| G+C content | 0.48180 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913862 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
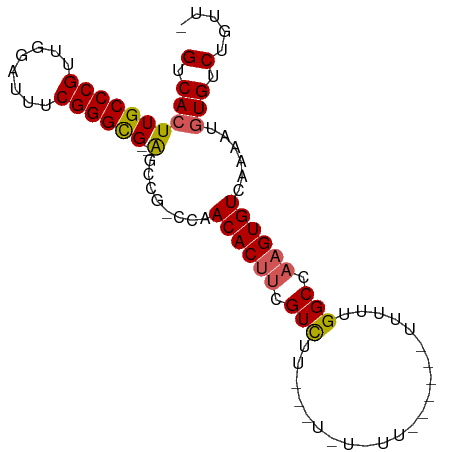
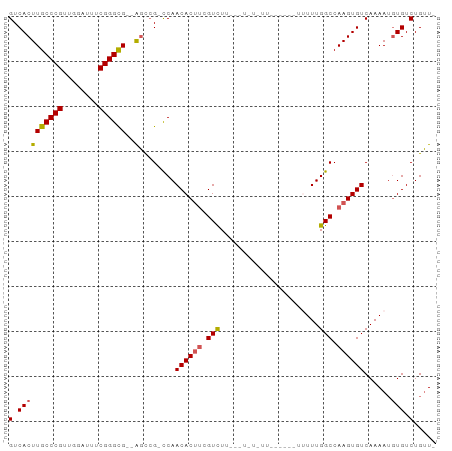
>dm3.chr3L 11921680 91 + 24543557 -AACAGACAUAUUUUGACACUUGGCCAAAAAAAGAAGAAAAAAGUCAAGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC -..((((.....)))).(((((((((.................(((((.((...)).))))-)))))--.(((((........)))))))))).. ( -21.92, z-score = -1.33, R) >droSim1.chr3L 11290520 89 + 22553184 -AACAGACACAUUUUGACACUUGGCCAAAAAAGAUGAAAAAAA--CAAGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC -......(((...(..((((((.(.(.................--...).).))))))..)-.....--.(((((........)))))..))).. ( -20.55, z-score = -0.97, R) >droYak2.chr3L 11966283 77 + 24197627 -AACAGACACAUUUUGACACUUGGCCAAA-----------------AAGACGAAGUGUUGGGCCGCCAACACCCGAAAUCCAACGGGCAAGUGAC -..((((.....)))).((((((.((...-----------------........(((((((....)))))))..(.....)...)).)))))).. ( -21.50, z-score = -1.66, R) >droEre2.scaffold_4784 11911592 74 + 25762168 -AACAGACACAUUUUGACACUUGGCCAAA-----------------AAGACGAAGUGUUGG-CGGCU--CGCCCGAAAUCCAACGGGCAAGUGAC -......(((...(..((((((.(.(...-----------------..).).))))))..)-.....--.(((((........)))))..))).. ( -22.20, z-score = -1.73, R) >droAna3.scaffold_13337 6173055 90 + 23293914 CACCAGACACUUUUUGACACUUGGCCAAAAAUACAGAAAUACAG--AAAACUGUGUGUUGA-CGAGC--CGCCCGAAAUCCAGCGGGCAAGUGAC .................(((((((((...((((((....)((((--....)))))))))..-.).))--)(((((........)))))))))).. ( -24.10, z-score = -2.36, R) >consensus _AACAGACACAUUUUGACACUUGGCCAAAAA______AA_A_A___AAGACGAAGUGUUGG_CGGCU__CGCCCGAAAUCCAACGGGCAAGUGAC .......(((...(..((((((.(..........................).))))))..).........(((((........)))))..))).. (-17.31 = -17.23 + -0.08)

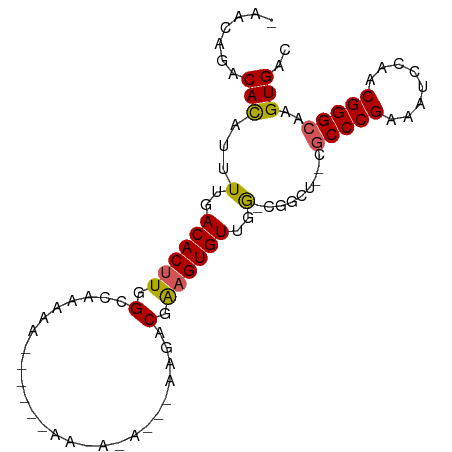
| Location | 11,921,680 – 11,921,771 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Shannon entropy | 0.36114 |
| G+C content | 0.48180 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.33 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826310 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
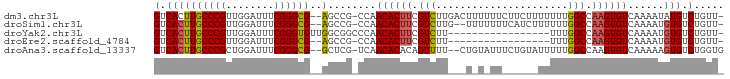
>dm3.chr3L 11921680 91 - 24543557 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCUUGACUUUUUUCUUCUUUUUUUGGCCAAGUGUCAAAAUAUGUCUGUU- ((..((((((((........))))))--))..)-)..((((((.(((......................))).))))))...............- ( -21.45, z-score = -1.21, R) >droSim1.chr3L 11290520 89 - 22553184 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCUUG--UUUUUUUCAUCUUUUUUGGCCAAGUGUCAAAAUGUGUCUGUU- ((..((((((((........))))))--))..)-)..((((((.(((...--.................))).))))))...............- ( -21.55, z-score = -1.13, R) >droYak2.chr3L 11966283 77 - 24197627 GUCACUUGCCCGUUGGAUUUCGGGUGUUGGCGGCCCAACACUUCGUCUU-----------------UUUGGCCAAGUGUCAAAAUGUGUCUGUU- (.((((((.(((..((((...(((((((((....))))))))).)))).-----------------..))).)))))).)..............- ( -26.10, z-score = -2.39, R) >droEre2.scaffold_4784 11911592 74 - 25762168 GUCACUUGCCCGUUGGAUUUCGGGCG--AGCCG-CCAACACUUCGUCUU-----------------UUUGGCCAAGUGUCAAAAUGUGUCUGUU- ((..((((((((........))))))--))..)-)..((((((.(((..-----------------...))).))))))...............- ( -23.20, z-score = -1.86, R) >droAna3.scaffold_13337 6173055 90 - 23293914 GUCACUUGCCCGCUGGAUUUCGGGCG--GCUCG-UCAACACACAGUUUU--CUGUAUUUCUGUAUUUUUGGCCAAGUGUCAAAAAGUGUCUGGUG .......(((.(((.(....).))))--)).(.-.((((((((((....--)))).........((((((((.....)))))))))))).))..) ( -23.30, z-score = -0.94, R) >consensus GUCACUUGCCCGUUGGAUUUCGGGCG__AGCCG_CCAACACUUCGUCUU___U_U_UU______UUUUUGGCCAAGUGUCAAAAUGUGUCUGUU_ (.(((..(((((........)))))............((((((.(((......................))).))))))......))).)..... (-17.05 = -17.33 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:59 2011