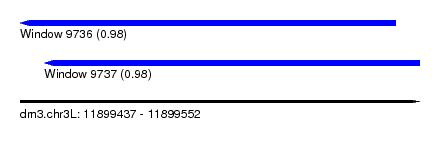
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,899,437 – 11,899,552 |
| Length | 115 |
| Max. P | 0.982770 |

| Location | 11,899,437 – 11,899,545 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.45 |
| Shannon entropy | 0.53201 |
| G+C content | 0.40910 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -11.75 |
| Energy contribution | -11.42 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11899437 108 - 24543557 ---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU------GCGC---AUGAUAUGGCAACCCUUUCCGGUGCU ---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..------))))---.))))..))).(((......)))... ( -30.90, z-score = -1.97, R) >droPer1.super_9 182809 91 - 3637205 ------------------CGUCCG-AGCAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACACGGAGAUUUAUUGCCCU------GCGC---AUGAUAUGGCAACCCUUUGGGCCCU- ------------------.(((((-((((((((....)))))))))......((((((........))))))..(((((.(------(...---....)).)))))......))))...- ( -22.40, z-score = -1.87, R) >dp4.chrXR_group6 1887117 91 - 13314419 ------------------CGUCCG-AGCAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACACGGAGAUUUAUUGCCCU------GCGC---AUGAUAUGGCAACCCUUUGGGCCCG- ------------------.(((((-((((((((....)))))))))......((((((........))))))..(((((.(------(...---....)).)))))......))))...- ( -22.40, z-score = -1.85, R) >droAna3.scaffold_13337 6153058 109 - 23293914 CGUCGCUGUGUCAGCUGGCCCUGG-AACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU------GCGC---AUGAUAUGGCAACCCUUUCAAGCCC- .((.(((((((((((.(((..(((-((((((((....)))))))))))....((((((........))))))....)))..------..))---.))))))))).))............- ( -32.90, z-score = -3.38, R) >droEre2.scaffold_4784 11889231 106 - 25762168 ---CGCUGCGUCAGCUGGCUCU-GUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU------GCGC---AUGAUGUGGCAACCCUUU-CAGUACU ---.((..(((((((.(((...-..((((((((....)))))))).......((((((........))))))....)))..------..))---.)))))..))........-....... ( -28.60, z-score = -2.47, R) >droYak2.chr3L 11942484 107 - 24197627 ---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU------GCGC---AUGAUAUGGCAACCCUUU-CAGUACU ---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..------))))---.))))..)))........-....... ( -29.40, z-score = -2.20, R) >droSec1.super_0 4087616 108 - 21120651 ---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCCU------GCGC---AUGAUAUGGCAACCCUUUUCGGUGCU ---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..------))))---.))))..))).(((......)))... ( -30.80, z-score = -1.93, R) >droSim1.chr3L 11267944 108 - 22553184 ---CGCUGCGUCAGUUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU------GCGC---AUGAUAUGGCAACCCUUUUCGGUGCU ---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..------))))---.))))..))).(((......)))... ( -30.90, z-score = -2.21, R) >droWil1.scaffold_180949 550153 108 + 6375548 UUUUUCCUUGUCAGUUGGCCUGAGAAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGAAGAUUUAUUGCUUG------GCGCCUGAUGAUAUGGCAACCCUUUCU------ .....((.(((((...((((..((.((((((((....)))))))).((((..((((((........)))))).))))))..------).)))...))))).))...........------ ( -26.60, z-score = -3.00, R) >droVir3.scaffold_13049 16944380 102 - 25233164 -----UGCUGUGUACUGGCCGCUAAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCUCUU------CGC---AUGGCAUGGCAGGCCUUUUAUA---- -----((((((((....((.....(((((((((....)))))))))((((..((((((........)))))).)))).....------.))---...))))))))...........---- ( -24.00, z-score = -2.09, R) >droMoj3.scaffold_6680 5026649 102 + 24764193 -----UGUGGCGCCUUGGCCCUUGAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCCUCU------CGC---ACGGGUCGACAAGCCUUUUACG---- -----...(((...(((((((((((((((((((....))))))))))))...((((((........))))))...(((....------.))---).)))))))...))).......---- ( -32.40, z-score = -5.22, R) >droGri2.scaffold_15110 9349361 108 + 24565398 -----UACUGCAUGCUGGGCUCCAAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCCCUUGCCUUGCGC---AUGGGUCGAGAAGUCCUUUAUA---- -----.....(((((.((((....(((((((((....)))))))))((((..((((((........)))))).)))).....))))...))---)))((.(.....).))......---- ( -26.40, z-score = -2.37, R) >consensus ___CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUU______GCGC___AUGAUAUGGCAACCCUUUUCGGU_C_ .........................((((((((....)))))))).......((((((........))))))..(((((......................))))).............. (-11.75 = -11.42 + -0.32)
| Location | 11,899,444 – 11,899,552 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.48971 |
| G+C content | 0.40264 |
| Mean single sequence MFE | -28.07 |
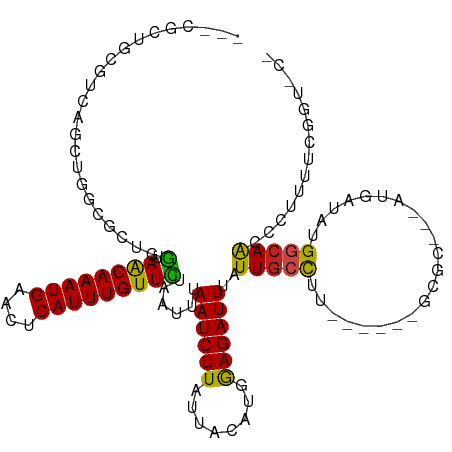
| Consensus MFE | -12.00 |
| Energy contribution | -11.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11899444 108 - 24543557 ACUUUCG---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC------AUGAUAUGGCAACCCUUUC .......---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..))))------.))))..)))......... ( -29.40, z-score = -2.42, R) >droPer1.super_9 182816 91 - 3637205 ------------------GCAUUCGCGUCCG-AGCAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACACGGAGAUUUAUUGCCCUGCGC------AUGAUAUGGCAACCCUUU- ------------------.(((.(((....(-((((((((....)))))))))((((..((((((........)))))).))))....))).------)))....(....).....- ( -20.20, z-score = -1.70, R) >dp4.chrXR_group6 1887124 91 - 13314419 ------------------GCAUUCGCGUCCG-AGCAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACACGGAGAUUUAUUGCCCUGCGC------AUGAUAUGGCAACCCUUU- ------------------.(((.(((....(-((((((((....)))))))))((((..((((((........)))))).))))....))).------)))....(....).....- ( -20.20, z-score = -1.70, R) >droAna3.scaffold_13337 6153065 109 - 23293914 ACUUUCGCGUCGCUGUGUCAGCUGGCCCUGG-AACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC------AUGAUAUGGCAACCCUUU- ........((.(((((((((((.(((..(((-((((((((....)))))))))))....((((((........))))))....)))....))------.))))))))).)).....- ( -32.90, z-score = -3.74, R) >droEre2.scaffold_4784 11889238 106 - 25762168 ACUUUCG---CGCUGCGUCAGCUGGCUCU-GUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC------AUGAUGUGGCAACCCUUU- .......---.((..(((((((.(((...-..((((((((....)))))))).......((((((........))))))....)))....))------.)))))..))........- ( -28.60, z-score = -2.67, R) >droYak2.chr3L 11942491 107 - 24197627 ACUUUCG---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC------AUGAUAUGGCAACCCUUU- .......---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..))))------.))))..)))........- ( -29.40, z-score = -2.35, R) >droSec1.super_0 4087623 108 - 21120651 ACUUUCG---CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCCUGCGC------AUGAUAUGGCAACCCUUUU .......---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..))))------.))))..)))......... ( -29.30, z-score = -2.27, R) >droSim1.chr3L 11267951 108 - 22553184 ACUUUCG---CGCUGCGUCAGUUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC------AUGAUAUGGCAACCCUUUU .......---.(((..((((....((((.(((((((((((....)))))))........((((((........))))))...))))..))))------.))))..)))......... ( -29.40, z-score = -2.50, R) >droWil1.scaffold_180949 550155 112 + 6375548 -AUUUCGUUUUUCCUUGUCAGUUGGCCUGAGAAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGAAGAUUUAUUGCUUGGCGCCU---GAUGAUAUGGCAACCCUUU- -.....(((...((.(((((...((((..((.((((((((....)))))))).((((..((((((........)))))).))))))..).))).---..))))).)).))).....- ( -26.90, z-score = -2.75, R) >droVir3.scaffold_13049 16944384 104 - 25233164 ---CUCU---CAUGCUGUGUACUGGCCGCUAAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCUCUUCGC------AUGGCAUGGCAGGCCUUU- ---((.(---((((((((((...........(((((((((....)))))))))((((..((((((........)))))).))))......))------))))))))).))......- ( -27.30, z-score = -3.33, R) >droMoj3.scaffold_6680 5026653 105 + 24764193 --AUUCG---CGUGUGGCGCCUUGGCCCUUGAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCCUCUCGC------ACGGGUCGACAAGCCUUU- --.....---.....(((...(((((((((((((((((((....))))))))))))...((((((........))))))...(((.....))------).)))))))...)))...- ( -32.40, z-score = -4.30, R) >droGri2.scaffold_15110 9349365 111 + 24565398 ----ACGUG-CUUACUGCAUGCUGGGCUCCAAAACAAAUGAACUCAUUUGUUUCAAUUUAAUCUUAUUACUCAAAGAUUUAUUGCCCUUGCCUUGCGCAUGGGUCGAGAAGUCCUU- ----.((((-(.....)))))..(((((.(.(((((((((....)))))))))((((..((((((........)))))).))))(((.(((.....))).)))....).)))))..- ( -30.80, z-score = -3.05, R) >consensus _CUUUCG___CGCUGCGUCAGCUGGCGCUGGUAACAAAUGAACUCAUUUGUUCUAAUUUAAUCUUAUUACAUGGAGAUUUAUUGCCUUGCGC______AUGAUAUGGCAACCCUUU_ .......................(((......((((((((....)))))))).......((((((........))))))....)))............................... (-12.00 = -11.78 + -0.22)

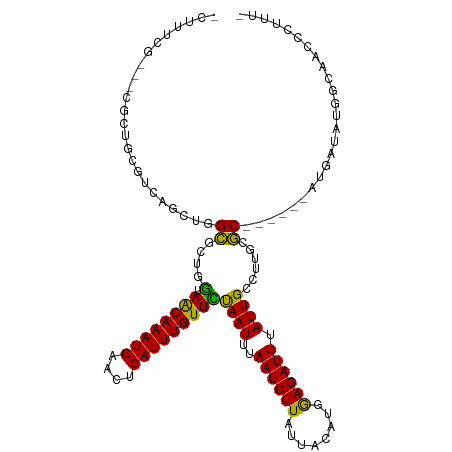
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:56 2011