
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,889,818 – 11,889,919 |
| Length | 101 |
| Max. P | 0.518458 |

| Location | 11,889,818 – 11,889,919 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Shannon entropy | 0.35269 |
| G+C content | 0.49580 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
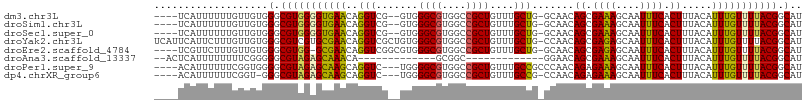
>dm3.chr3L 11889818 101 - 24543557 ----UCAUUUUUUGUUGUGGGCGUGGGGUGAACAGGUCG--GUGGGCGUGGCCGCUGUUUGCUG-GCAACAGCGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----........((((((((((((((((((((..(((((--(....).)))))(((.(((((((-....))))))))))...))))))))))....).))))))))). ( -36.20, z-score = -2.63, R) >droSim1.chr3L 11258423 101 - 22553184 ----UCAUUUUUUGUUGUGGGCGUGGGGUGAACAGGUCG--GUGGGCGUGGCCGCUGUUUGCUG-GCAACAGCGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----........((((((((((((((((((((..(((((--(....).)))))(((.(((((((-....))))))))))...))))))))))....).))))))))). ( -36.20, z-score = -2.63, R) >droSec1.super_0 4077991 101 - 21120651 ----UCAUUUUUUGUUGUGGGCGUGGGGUGAACAGGUCG--GUGGGCGUGGCCGCUGUUUGCUG-GCAACAGCGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----........((((((((((((((((((((..(((((--(....).)))))(((.(((((((-....))))))))))...))))))))))....).))))))))). ( -36.20, z-score = -2.63, R) >droYak2.chr3L 11931492 107 - 24197627 UCAUUCAUUCUUUGUUGUGGGCGUCGUGCGAACAGGUCGCUGUGGGCGUGGCCGCUGUUUGCUG-CCAACAGCGAGAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ...(((((........))))).((((((.((((((((.(((((.....((((.((.....)).)-))))))))((((....)))).......)))))))))))))).. ( -31.90, z-score = 0.09, R) >droEre2.scaffold_4784 11879688 102 - 25762168 ----UCGUUCUUUGUUGUGGGCGUGG-GCGAACAGGUCGGCGUGGGCGUGGCCGCUGUUUGCUG-GCAACAGCGAGAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----..((((((((((((.((((..(-(((..((.(((......))).))..))))...)))).-))))))..))))))................(((......))). ( -33.30, z-score = -0.32, R) >droAna3.scaffold_13337 6145140 80 - 23293914 --ACUCAUUUUUUUUCGGGGGCGUAGAGCAAACA-------------GCGGC-------------GGAACAGCGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU --.(((..........)))(.(((((((((((..-------------..((.-------------((((..((....))..)))).)).....))))))))))).).. ( -22.10, z-score = -3.75, R) >droPer1.super_9 174033 101 - 3637205 ----ACAUUUUUUCGGUGGGGCGUAGAGCAAGCAGGUC---UGGGGCGUGGCCGCUGUUUGCCGCCCAACAGAGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----.((((.....)))).(.(((((((((((....((---((.((.(((((........)))))))..))))((((....))))........))))))))))).).. ( -31.40, z-score = -0.83, R) >dp4.chrXR_group6 1878449 99 - 13314419 ----ACAUUUUUUCGGU-GGGCGUAGAGCAAGCAGGUC---UGGGGCGUGGCCGCUGUUUGCCG-CCAACAGAGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ----.((((.....)))-)(.(((((((((((....((---((..(.(((((........))))-))..))))((((....))))........))))))))))).).. ( -31.40, z-score = -1.32, R) >consensus ____UCAUUUUUUGUUGUGGGCGUGGAGCGAACAGGUCG__GUGGGCGUGGCCGCUGUUUGCUG_GCAACAGCGAAAGCAAUUUCACUUUACAUUUGUUUUACGGCAU ...................(.(((((((((((((.(((......))).))....(((((........))))).((((....))))........))))))))))).).. (-18.31 = -18.36 + 0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:52 2011