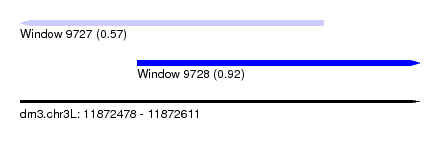
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,872,478 – 11,872,611 |
| Length | 133 |
| Max. P | 0.924468 |

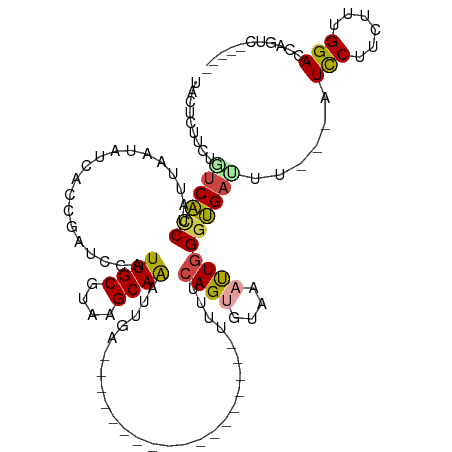
| Location | 11,872,478 – 11,872,579 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.56663 |
| G+C content | 0.42325 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11872478 101 - 24543557 ------------------UACACUGAAAAUCAAUUUGCUUACGCAAUGGAUCGGUGAUAUUAAUAGGUGACAGAAGAGUAUUCAGCGAAAGCUUACAGUCCACCUUAAAGGACUCUUCU- ------------------..((((((...((...((((....))))..)))))))).........(....)(((((((((...(((....)))))).((((........))))))))))- ( -29.70, z-score = -3.68, R) >droSim1.chr3L 11241569 101 - 22553184 ------------------UACACUGAAAAUUAAUUUGCUUACGCAAUGGAUCGGUGAUAUUAAUAGGUGACAGAAGGGUAUUCAGCGGAAGCUUACAGUCCACCUUAAAGGACUCUUCU- ------------------..((((((...((...((((....))))..)))))))).........(....)(((((.(((...(((....))))))(((((........))))))))))- ( -26.90, z-score = -2.40, R) >droSec1.super_0 4061249 101 - 21120651 ------------------UACACUGAAAAUCAAUUUGCUUACGCAAUGGAUCGAUGAUAUUGAUAGGUGACAGAAGGGUAUUCAGCGAAAGCUUACAGUCCACCUUAAAGGACUCUUCU- ------------------..((((.....(((..((((....)))))))((((((...)))))).))))..(((((.(((...(((....))))))(((((........))))))))))- ( -24.20, z-score = -1.66, R) >droYak2.chr3L 11914722 119 - 24197627 CACACUGAAAAUCUAGCAGGUGCUUGAUCUCAAUUUGCUUACGCAAUGGAUCGGUGAUGUUAAUUGGUGACAGAAGAGUGUGCAGCGGAAGCUUACAGUCCACCUUAAAGGACUGUUCU- ((((((.......(((((..((((.(((((....((((....)))).))))))))).))))).(((....)))...)))))).(((....))).(((((((........)))))))...- ( -38.30, z-score = -2.22, R) >droEre2.scaffold_4784 11863139 119 - 25762168 CACACUGAAAAUCUAACAGGUGCUUAAUCUCAAUUUGCUUACGCAAUGGAUCGGUGAUAUUAAUUGGUGAUAGAAGAGUAUGCAGCGGUAGCUUACAGUCCACCUUAAGGGCCUCUUCA- ..((((((...((((..((((.....))))....((((....))))))))))))))................((((((.....(((....)))....((((.......)))))))))).- ( -26.30, z-score = 0.35, R) >droAna3.scaffold_13337 6129152 117 - 23293914 UACGCUGGAACGACGGAAGGUCCUUUACCUCACUCUGCUUGCGCAAACUCUCGAUUAUGUCGAUGGAGGGAAAAAGAUUAUUGAGUG---GCCAGGUGGGUACUUCAAAGGAUGGCGUCA .((((((...(....((((..(((..((((((((((((....)))..(((((.(((.....))).)))))............)))))---...)))))))..))))...)..)))))).. ( -33.80, z-score = -0.36, R) >consensus __________________GACACUGAAAAUCAAUUUGCUUACGCAAUGGAUCGGUGAUAUUAAUAGGUGACAGAAGAGUAUUCAGCGGAAGCUUACAGUCCACCUUAAAGGACUCUUCU_ ..................................((((....))))...(((.((.......)).)))....(((((((....(((....))).........((.....))))))))).. (-11.06 = -11.20 + 0.14)

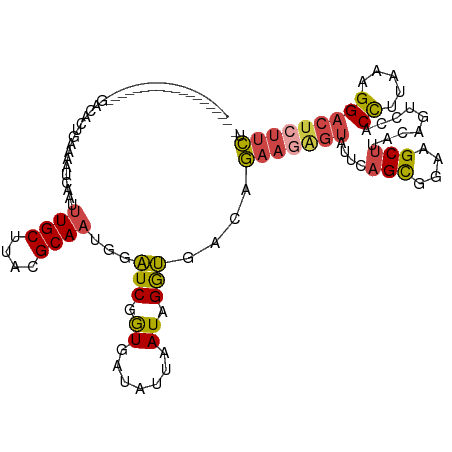
| Location | 11,872,517 – 11,872,611 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.11 |
| Shannon entropy | 0.52363 |
| G+C content | 0.40667 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -12.33 |
| Energy contribution | -14.17 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11872517 94 + 24543557 UACUCUUCUGUCACCUAUUAAUAUCACCGAUCCAUUGCGUAAGCAAAUUGA------------------UUUUCAGUGUAAAUUGGGUGAUUU---AUCCUUCUUUGGACCAGUC----- .......((((((((((.......(((.((((..((((....))))...))------------------))....))).....)))))))...---.(((......))).)))..----- ( -21.30, z-score = -2.31, R) >droSim1.chr3L 11241608 94 + 22553184 UACCCUUCUGUCACCUAUUAAUAUCACCGAUCCAUUGCGUAAGCAAAUUAA------------------UUUUCAGUGUAAAUUGGAUGACUU---AUCCUGCCUUGGACCAGUC----- .............................((((.((((....))))...((------------------(((.......)))))))))((((.---.(((......)))..))))----- ( -15.70, z-score = -0.79, R) >droSec1.super_0 4061288 94 + 21120651 UACCCUUCUGUCACCUAUCAAUAUCAUCGAUCCAUUGCGUAAGCAAAUUGA------------------UUUUCAGUGUAAUUUGGGUGACUU---AUCCUUCUUUGGACCAGUC----- .........((((((((.((.....((((((...((((....)))))))))------------------)......)).....))))))))..---.(((......)))......----- ( -22.60, z-score = -2.64, R) >droYak2.chr3L 11914761 117 + 24197627 CACUCUUCUGUCACCAAUUAACAUCACCGAUCCAUUGCGUAAGCAAAUUGAGAUCAAGCACCUGCUAGAUUUUCAGUGUGGAUUGGGUGAAUU---AUCCUUCUUUGGACCAUUUGAACA ..........(((..........(((((((((((((((....))))(((((((((.(((....))).)).))))))).)))))).)))))...---.(((......))).....)))... ( -32.10, z-score = -2.42, R) >droEre2.scaffold_4784 11863178 112 + 25762168 UACUCUUCUAUCACCAAUUAAUAUCACCGAUCCAUUGCGUAAGCAAAUUGAGAUUAAGCACCUGUUAGAUUUUCAGUGUGGAUUGGGUGAUUU---AUCCUUUUUUGGACCAUUU----- .............((((..((.((((((((((((((((....))))(((((((((.(((....))).)).))))))).)))))).))))))..---....))..)))).......----- ( -27.90, z-score = -2.51, R) >droAna3.scaffold_13337 6129189 115 + 23293914 UAAUCUUUUUCCCUCCAUCGACAUAAUCGAGAGUUUGCGCAAGCAGAGUGAGGUAAAGGACCUUCCGUCGUUCCAGCGUAGUCUCGGGGAUUUUCAAUUCGCCGCCGGACUGGAC----- ...((((((..(((((.((((.....))))...(((((....)))))).)))).))))))...(((..(((....))).((((((((.((........)).)))..)))))))).----- ( -33.90, z-score = -0.64, R) >consensus UACUCUUCUGUCACCUAUUAAUAUCACCGAUCCAUUGCGUAAGCAAAUUGA__________________UUUUCAGUGUAAAUUGGGUGAUUU___AUCCUUCUUUGGACCAGUC_____ .........((((((...................((((....))))...........................((((....))))))))))......(((......)))........... (-12.33 = -14.17 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:48 2011