| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,857,600 – 11,857,705 |
| Length | 105 |
| Max. P | 0.999645 |

| Location | 11,857,600 – 11,857,705 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Shannon entropy | 0.34832 |
| G+C content | 0.46176 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -24.01 |
| Energy contribution | -26.82 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
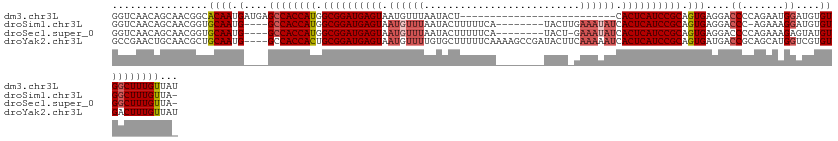
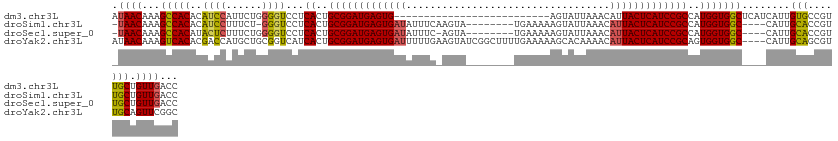
>dm3.chr3L 11857600 105 + 24543557 GGUCAACAGCAACGGCACAAUGAUGAGCCACCAUGGCGGAUGAGUAAUGUUUAAUACU--------------------------CACUCAUCCGCAGUGAGGACCCCAGAAUGGAUGUGUGGCUUUGUUAU ....((((((....))........((((((((((.((((((((((...((.....)).--------------------------.))))))))))..........((.....))))).))))))))))).. ( -34.70, z-score = -1.77, R) >droSim1.chr3L 11227231 117 + 22553184 GGUCAACAGCAACGGUGCAAUG----GCCACCAUGGCGGAUGAGUAAUGUUUAAUACUUUUUCA--------UACUUGAAAUAUCACUCAUCCGCAGUGAGGACCC-AGAAAGGAUGUGUGGCUUUGUUA- ....(((((((....)))...(----((((((((.((((((((((.((((((............--------......)))))).)))))))))).........((-.....))))).)))))).)))).- ( -36.07, z-score = -1.79, R) >droSec1.super_0 4047782 117 + 21120651 GGUCAACAGCAACGGUGCAAUG----GCCACCAUGGCGGAUGAGUAAUGUUUAAUACUUUUUCA--------UACU-GAAAUAUCACUCAUCCGCAGUGAGGACCCCAGAAAGAGUAUGUGGCUUUGUUA- ....(((((((....)))...(----((((((((.((((((((((.((((((............--------....-.)))))).)))))))))).))).((...))...........)))))).)))).- ( -34.63, z-score = -1.60, R) >droYak2.chr3L 11900738 127 + 24197627 GCCGAACUGCAACGCUGCAAUG----GCCACCACUGCGGAUGAGUAAUGUUUUGUGCUUUUUCAAAAGCCGAUACUUCAAAAAUCACUCAUCCGCAGUGAUGACCGCAGCAUGGUCGUGUGACUUUGUUAU ((((...((((....)))).))----))(((((((((((((((((.((((.(((.(((((....)))))))).)).......)).))))))))))))))(((((((.....)))))))))).......... ( -46.91, z-score = -4.18, R) >consensus GGUCAACAGCAACGGUGCAAUG____GCCACCAUGGCGGAUGAGUAAUGUUUAAUACUUUUUCA________UACU_GAAAUAUCACUCAUCCGCAGUGAGGACCCCAGAAAGGAUGUGUGGCUUUGUUA_ ....(((((((....)))........((((((((.((((((((((.((((((..........................)))))).)))))))))).)))....((.......))....)))))..)))).. (-24.01 = -26.82 + 2.81)
| Location | 11,857,600 – 11,857,705 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Shannon entropy | 0.34832 |
| G+C content | 0.46176 |
| Mean single sequence MFE | -43.41 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.40 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
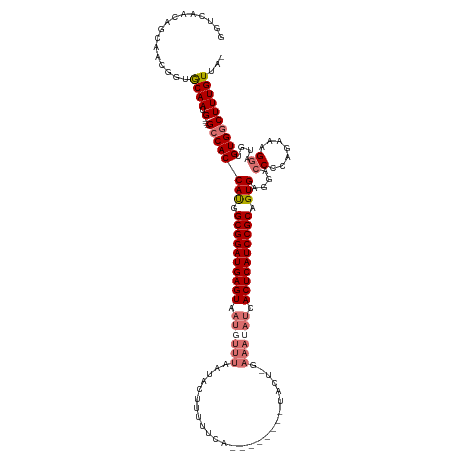
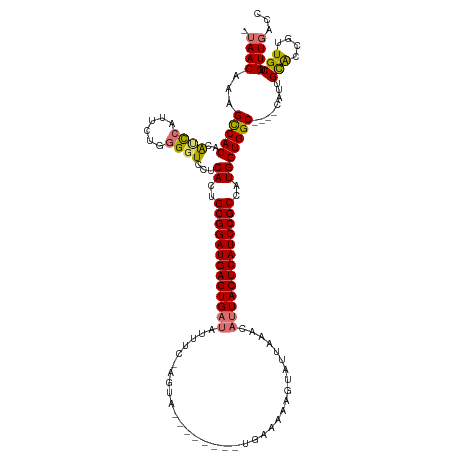
>dm3.chr3L 11857600 105 - 24543557 AUAACAAAGCCACACAUCCAUUCUGGGGUCCUCACUGCGGAUGAGUG--------------------------AGUAUUAAACAUUACUCAUCCGCCAUGGUGGCUCAUCAUUGUGCCGUUGCUGUUGACC .(((((.(((..((((.........(((((.(((..(((((((((((--------------------------(((.....)).))))))))))))..))).))))).....))))..)))..)))))... ( -36.54, z-score = -2.89, R) >droSim1.chr3L 11227231 117 - 22553184 -UAACAAAGCCACACAUCCUUUCU-GGGUCCUCACUGCGGAUGAGUGAUAUUUCAAGUA--------UGAAAAAGUAUUAAACAUUACUCAUCCGCCAUGGUGGC----CAUUGCACCGUUGCUGUUGACC -(((((..(((((..((((.....-))))...((..(((((((((((((.(((((....--------)))))..((.....)))))))))))))))..)))))))----....(((....))))))))... ( -38.20, z-score = -3.70, R) >droSec1.super_0 4047782 117 - 21120651 -UAACAAAGCCACAUACUCUUUCUGGGGUCCUCACUGCGGAUGAGUGAUAUUUC-AGUA--------UGAAAAAGUAUUAAACAUUACUCAUCCGCCAUGGUGGC----CAUUGCACCGUUGCUGUUGACC -(((((..(((((..(((((....)))))...((..(((((((((((((.(((.-((((--------(......)))))))).)))))))))))))..)))))))----....(((....))))))))... ( -39.80, z-score = -3.84, R) >droYak2.chr3L 11900738 127 - 24197627 AUAACAAAGUCACACGACCAUGCUGCGGUCAUCACUGCGGAUGAGUGAUUUUUGAAGUAUCGGCUUUUGAAAAAGCACAAAACAUUACUCAUCCGCAGUGGUGGC----CAUUGCAGCGUUGCAGUUCGGC ........(((..((..(.(((((((((((((((((((((((((((((((((((........(((((....))))).))))).))))))))))))))))))))))----)...))))))).)..))..))) ( -59.10, z-score = -7.18, R) >consensus _UAACAAAGCCACACAUCCAUUCUGGGGUCCUCACUGCGGAUGAGUGAUAUUUC_AGUA________UGAAAAAGUAUUAAACAUUACUCAUCCGCCAUGGUGGC____CAUUGCACCGUUGCUGUUGACC .((((...(((((..((((......))))...((..(((((((((((((..................................)))))))))))))..)))))))........(((....))).))))... (-27.67 = -27.86 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:45 2011