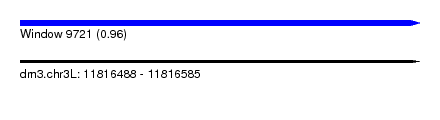
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,816,488 – 11,816,585 |
| Length | 97 |
| Max. P | 0.957621 |

| Location | 11,816,488 – 11,816,585 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 58.99 |
| Shannon entropy | 0.89987 |
| G+C content | 0.52297 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.14 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11816488 97 + 24543557 ----GAUAGAAGCUCCCAUGGGAGUUGAGUUCAUCACGUCAUCAGGUAGUCGAGGACAUGUGG-------CUGUCACAUCUCGACGGACCUGCACUCAGGGUCUUUAA ----....((((..(((.((((.((.((.....)))).....(((((.((((((...(((((.-------....)))))))))))..)))))..))))))).)))).. ( -31.30, z-score = -0.81, R) >droSim1.chr3L 11181953 97 + 22553184 ----GAGAGAAGCUCCCAUGGGAGUUGAGUCCAUCACGACAUCAGGUAGUCGAGGACAUGUGG-------CUGUCACAUCUCGACGGACCUGCACUCAGGGUCUUUAA ----..(((.((((((....))))))..(((......)))..(((((.((((((...(((((.-------....)))))))))))..)))))..)))........... ( -33.60, z-score = -0.90, R) >droSec1.super_0 4001898 97 + 21120651 ----GAGAGAAGCUCCCAUGGGAGUUGAGUCCAUCACGUCAUCAGGUAGUCGAGCACAUGUGG-------CUGUCACAUCUCGACGGACCUGCACUCAGGGUCUUUAA ----.(((((((((((....))))))((((............(((((.((((((...(((((.-------....)))))))))))..))))).))))....))))).. ( -32.92, z-score = -0.96, R) >droYak2.chr3L 11849604 96 + 24197627 ----GAGAGAAGCUCCUGCGGGAGUCGAGUCCAUCACGUCAUCAGGUAGUCGAGAC-AUGUGG-------CUGCCACAUCUCGACGGACCUGCACUCAGGGUCUUUAA ----....((((..((((.((((......)))..........(((((.(((((((.-.(((((-------...))))))))))))..)))))..).))))..)))).. ( -35.30, z-score = -1.15, R) >droEre2.scaffold_4784 11799589 97 + 25762168 ----GAAAGAUGCUCCUCCGGGAGUCGAAUCCAUCACGUCAUCAGGUAGUCGAGACCAUGUGG-------CUGUCACAUCUCGACGGACCUGCACUCAGGGUCUUUAA ----.(((((((((((....))))).................(((((.(((((((...((((.-------....)))))))))))..)))))........)))))).. ( -31.30, z-score = -0.81, R) >droAna3.scaffold_13337 17230448 92 - 23293914 -----AAUGGAACUCCUUUGGGAGCAGAUUUUGCCACUGCAUCAGGUAGUCGAG----UGCGA-------CAGCAAGCACUCGGCGGACCUUCACUCAGGGUCUUAAA -----..........(((((((.((((.........))))...((((.((((((----(((..-------......)))))))))..))))...)))))))....... ( -32.50, z-score = -2.00, R) >dp4.chrXR_group8 4294892 90 + 9212921 -----ACGCCUGCUCCC-CGCGAGU--AGUGUGCCACAGCGUUAGGAAGUCGAU--CAUGUGG-------UAAC-AUUGAUCGGCGGACCUGCACUCAGGGUCUUAAA -----.(((((((((..-...))))--)).)))........((((((.((((((--(((((..-------..))-).))))))))...((((....)))).)))))). ( -31.30, z-score = -1.13, R) >droPer1.super_29 528854 90 + 1099123 -----ACGCCUGCUCCC-CGCGAGU--AGUGUGCCACAGCGUUAGGAAGUCGAU--CAUGUGG-------UAAC-AUUGAUCGGCGGACCUGCACUCAGGGUCUUAAA -----.(((((((((..-...))))--)).)))........((((((.((((((--(((((..-------..))-).))))))))...((((....)))).)))))). ( -31.30, z-score = -1.13, R) >droMoj3.scaffold_6680 13393451 93 + 24764193 ---------AAAUCCAAAUCUGG----AA--GGCUUCAGCGUCAGGUAGUCGCAUGUAUAUGUUCUCUUUCACAUACAUAGCGGCGGACCUGCACUCAGGGUCUUUAA ---------...((((....)))----)(--(((((.((.(.(((((.(((((((((((.((........)).)))))).)))))..)))))).))..)))))).... ( -31.00, z-score = -3.91, R) >droVir3.scaffold_13049 11303485 91 - 25233164 ------AUAUGAUUCCCAUUGGG----AACUGGCUUCAGCGUUAGGCAGCCGUAUUUGUGC-------UCAAACCACAUUACGGCGGACCUGCACUCAGGGUCUUAAA ------......(((((...)))----))(((((......)))))...((((((..((((.-------......)))).))))))((((((.......)))))).... ( -28.30, z-score = -1.51, R) >droGri2.scaffold_15110 15310611 97 + 24565398 AAGAGAUGAAGAGUCAUCCCUGG----CUGGCUACACAGUGUUAGGCAGUCGAAUGUGUGUGU-----GUAUACCACAU--CGGCGGACCUGCACUCAGGGUCUUAAA (((((((((....)))))(((((----(((......))))(((((((.(((((.((((.(((.-----...))))))))--)))).).)))).)).)))).))))... ( -31.80, z-score = -0.95, R) >anoGam1.chr3L 21457879 90 - 41284009 ----------------UGCACGAAACGCGUGCA-CACACUGUCCGGGCACGGUAUUCUCAGGAAGCUUCCCUCGUGUGCGGAAGCGGACCUGCACUCAGGGUCUUAA- ----------------((((((.....))))))-....(((.(((....)))......)))...(((((((......).))))))((((((.......))))))...- ( -30.10, z-score = -0.26, R) >triCas2.ChLG4 6905670 84 - 13894384 ----UAAAUUAAUUCAUUUAUGCUAUAAACGGUUCAUAG-AUCAGGGAGCUGGG------------------GUUAUCCC-UGGCGGACCUGUACCCAGGGUCUUAAA ----..........................((..(....-..((((..((..((------------------(....)))-..))...))))......)..))..... ( -21.60, z-score = -0.30, R) >consensus _____AAAGAAGCUCCCAUGGGAGU_GAGUGCAUCACAGCAUCAGGUAGUCGAG__CAUGUGG_______CUGCCACAUCUCGGCGGACCUGCACUCAGGGUCUUAAA ................................................((((((.........................))))))((((((.......)))))).... (-11.65 = -11.14 + -0.51)

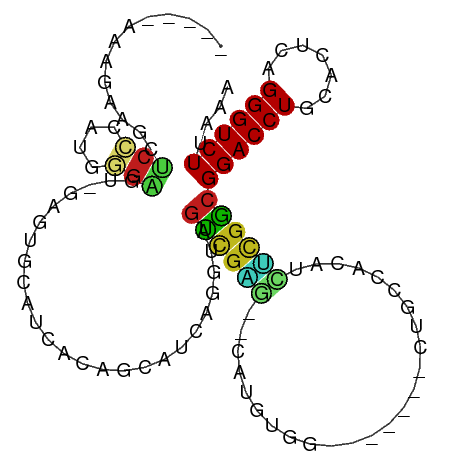
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:43 2011