| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,775,974 – 11,776,073 |
| Length | 99 |
| Max. P | 0.542370 |

| Location | 11,775,974 – 11,776,073 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 56.91 |
| Shannon entropy | 0.91881 |
| G+C content | 0.43227 |
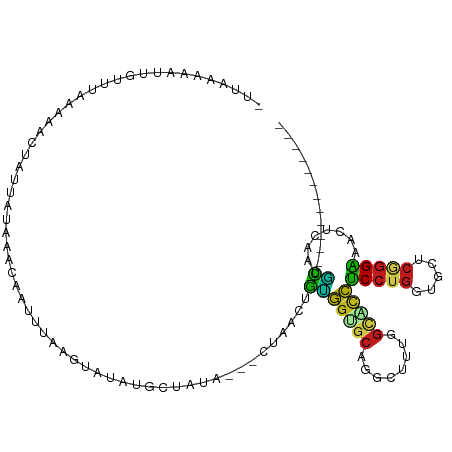
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -8.90 |
| Energy contribution | -8.72 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11775974 99 + 24543557 -UACAAGAUUACAUUAGAACUAUUUUCAACCAUUUGUAUAUGUGCUGUA---CUAACUGUGGUGCAGGUUUUGGCACCUCCUGGUGCUCAGGAAACUCAAUGC------------ -..(((((((((((....((...............))..)))))(((((---(((....))))))))))))))(((((....)))))..((....))......------------ ( -23.26, z-score = -0.38, R) >droSim1.chr3L 11141493 99 + 22553184 -UGAAAGAUUUCAUCAGAACUAUUUUUAACCAUUUAUGUAUAUGCUGUA---CUAACUGUGGUGCAGGUUUUGGCACCUCCUGGUGCUCGGGAAACUCAAUGC------------ -.....((((((..((((((..........(((........)))(((((---(((....))))))))))))))(((((....)))))....)))).)).....------------ ( -24.90, z-score = -1.30, R) >droSec1.super_0 3960959 99 + 21120651 -UAAAAGAUUUCGUCAGAACUAUAUUUAACCAUUUAUGUAUAUGGUGUA---CUAACUGUGGUGCAGGUUUUGGCACCUCCUGGUGCUCGGGAAACUCAAUGC------------ -.....((((((..(((((((.......(((((........)))))(((---(((....)))))).)))))))(((((....)))))....)))).)).....------------ ( -27.60, z-score = -2.08, R) >droYak2.chr3L 11804987 99 + 24197627 -UAACAGAUGUUAUCAGAACUAUUAUUAAAAAUUGUAAUAUAUGCUAUA---CUAACUGUGGUGCAGGUUUUGGCGCCUCCUGGUGCUCGGGAAACUCUAUGC------------ -((((....)))).((((((((((((........)))))...(((..((---(.....)))..))))))))))(((((....)))))..((....))......------------ ( -21.40, z-score = -0.55, R) >droEre2.scaffold_4784 11753707 97 + 25762168 ---AGAACUUUUGUCAGAACUAUUAUAAACAAUUUGUAUAUAUGCUAUA---CUAACUGCGGUGCAGGUUUUGGCACCUCCUGGUGCUCAGGAAACUCUAUGC------------ ---(((.....(((((((((...............(((((.....))))---)...((((...)))))))))))))..(((((.....)))))...)))....------------ ( -21.90, z-score = -0.83, R) >droAna3.scaffold_13337 6029074 99 + 23293914 UCCAAAAUGUGUUUAAAUACGUUCCUAAUCUCGCUUAGUUUGGG--AAUA--CUUACUGUGGUGCUGGCUUUGGUAUCUCCUGAUGCUCGGGGAACUCAAUGC------------ .......((.((((......((((((((.((.....)).)))))--))).--........((((((......)))))).((((.....)))))))).))....------------ ( -20.70, z-score = -0.12, R) >dp4.chrXR_group8 4881391 100 - 9212921 ACCGAGA-CUGUUAGAAGACUGUGCCGAACUGGCCGAGUCGAGGCCACAA--CUCACUGCGGGGCUGGCUUGGGCACCUCCUGGUGCUCCGGGAACUCGAUGC------------ ..((((.-.............(((((.(((..(((..((.(((.......--)))...))..)))..).)).))))).((((((....)))))).))))....------------ ( -37.20, z-score = -0.31, R) >droPer1.super_29 533516 100 - 1099123 ACCGAGA-GUGUUAGAAGACUGUGCCGAACUGGCCGAGUCGAGGCAACAA--CUCACUGCGGGGCUGGCUUGGGCACCUCCUGGUGCUCCGGGAACUCGAUGC------------ .(((.((-(..((((.((...(((((.(((..(((..((.(((.......--)))...))..)))..).)).))))))).))))..))))))...........------------ ( -38.70, z-score = -0.71, R) >droWil1.scaffold_180949 3820708 85 - 6375548 -------------UAGAAAAGAAGAGUAAGAAUUUAAUUAACU--UCUA---CUCACUGUGGAGCAGGCUUGGGAGCCUCCUGAUGUUCGGGAAAUUGUAUAU------------ -------------..........(((((.(((.((....)).)--))))---)))........(((((((....))))(((((.....)))))...)))....------------ ( -17.80, z-score = -0.12, R) >droVir3.scaffold_13049 24000206 98 + 25233164 AAUGUAAAUCCAAUAAGUGCAU---CACUGUCAAUAAUU-CACGAUUAUAU-CGUACUGUGGUGCGGGCUUGGGCGCCUCCUGAUGCUCGGGGAACUCUAUGC------------ ........(((.......((((---(((.((........-.(((((...))-))))).)))))))(((((..((.....))..).))))..))).........------------ ( -24.70, z-score = -0.31, R) >droMoj3.scaffold_6680 18631494 101 + 24764193 AAUUUAAUUCGAUAAAAUUGAGCCACAAGUAAAAUAUUUCAAUGGUUAAG--CUUACUGUGGAGCAGGCUUAGGUAUCUCCUGAUGCUCAGGGAACUCAAUGC------------ .......(((((((...(((((((..(((((...)))))..........(--(((......)))).))))))).)))).((((.....)))))))........------------ ( -21.30, z-score = -0.32, R) >droGri2.scaffold_15110 14871031 99 - 24565398 UGUUUAAUUUGAGUAAAUAAAU---CAAUCGAAAUAAUU-GGUGGCAGAGAACUUACUGCGGUGCAGGCUUGGGCGUCUCCUGAUGCUCGGGAAACUCAAUGC------------ ........((((((......((---(.(((((.....))-))).((((........))))))).....((((((((((....))))))))))..))))))...------------ ( -26.20, z-score = -1.48, R) >apiMel3.Group1 18263688 111 - 25854376 -AUGUAGUUUGUUGUGUGAUAUCUAUUAAUAAUUCUUGUGUAUUCUGAUCCAAUGAAUUAAAAUCAAG---AGAUUCUACAUCAUAAUCACGUAAUUCUGUUGGGAAUAAUACUC -((((.(.((((.(((((..((((.....(((((((((..(......)..))).))))))........---))))..))))).)))).))))).(((((....)))))....... ( -15.32, z-score = 0.90, R) >consensus _UUAAAAAUUGUUUAAAAACUAUUAUAAACAAUUUAAGUAUAUGCUAUA___CUAACUGUGGUGCAGGCUUUGGCACCUCCUGGUGCUCGGGAAACUCAAUGC____________ ............................................................(((((........)))))(((((.....)))))...................... ( -8.90 = -8.72 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:38 2011