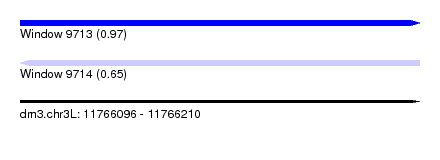
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,766,096 – 11,766,210 |
| Length | 114 |
| Max. P | 0.973879 |

| Location | 11,766,096 – 11,766,210 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.20 |
| Shannon entropy | 0.31201 |
| G+C content | 0.45127 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
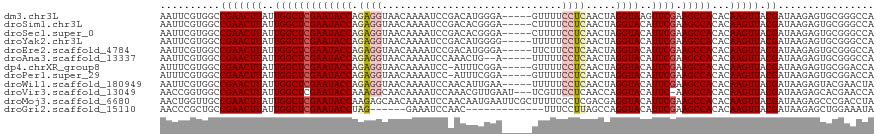
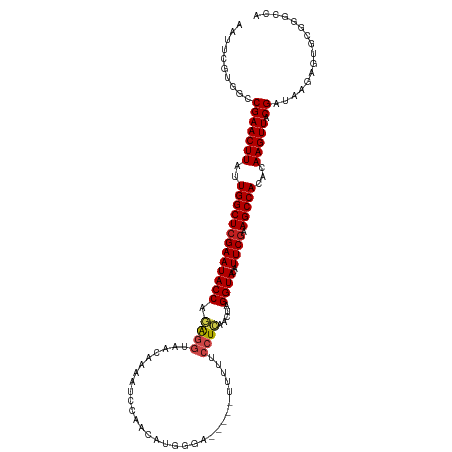
>dm3.chr3L 11766096 114 + 24543557 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAACUUACCUAGUUGAGGAAAAC-----UCCCAUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.((((..((((.....((((.(((.-----(((......))).)))....)))).)))))))))))))))))))).)))))...... ( -34.00, z-score = -2.32, R) >droSim1.chr3L 11131599 114 + 22553184 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAG-----UCCCGUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.(((..(((((.....((((.((((-----(((......)))))))....)))).)))))))))))))))))))).)))))...... ( -38.20, z-score = -3.10, R) >droSec1.super_0 3951119 114 + 21120651 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAG-----UCCCGUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.(((..(((((.....((((.((((-----(((......)))))))....)))).)))))))))))))))))))).)))))...... ( -38.20, z-score = -3.10, R) >droYak2.chr3L 11794901 114 + 24197627 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAA-----CCCCAUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.(((..(((((.....((((.((((-----.((......)).))))....)))).)))))))))))))))))))).)))))...... ( -33.60, z-score = -2.06, R) >droEre2.scaffold_4784 11743708 114 + 25762168 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAGAA-----UCCCAUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.(((..(((((.....((((.((((-----(((......)))))))....)))).)))))))))))))))))))).)))))...... ( -38.30, z-score = -3.23, R) >droAna3.scaffold_13337 6019502 112 + 23293914 UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAA-----U--CAGUUUGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU (((((...((.......))(((((((.(((((.(((..(((((.....((((.((((-----(--(......))))))....)))).)))))))))))))))))))).)))))...... ( -35.40, z-score = -2.87, R) >dp4.chrXR_group8 4870686 113 - 9212921 UGGUCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAC-----UCCGAAAU-GGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAAU (((.(((............(((((((.(((((.(((..(((((.....((((.(((.-----(((.....-))).)))....)))).))))))))))))))))))))))))))...... ( -33.00, z-score = -2.14, R) >droPer1.super_29 522786 113 - 1099123 UGGUCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAC-----UCCGAAAU-GGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAAU (((.(((............(((((((.(((((.(((..(((((.....((((.(((.-----(((.....-))).)))....)))).))))))))))))))))))))))))))...... ( -33.00, z-score = -2.14, R) >droWil1.scaffold_180949 3805278 114 - 6375548 UAGUUCGUACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAA-----UUCAAUGUUGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU .(((((((.......(((.(((((((.(((((.(((..(((((.....((((.((((-----((((....))))))))....)))).)))))))))))))))))))))))..))))))) ( -35.80, z-score = -3.84, R) >droVir3.scaffold_13049 23988145 115 + 25233164 UGGUUCGUGCUCUUAUCGUAACUUGUGUGGCUU-GAAUGUACCUGGUUGAGGAACGA---AUUCAACGUUUGGAUUUUGUUGCCUUUGGUAUUCGAGCCAAUAAGUUCGGCCACCGGUU .(((..(.((......((.(((((((.((((((-((..(((((.....(((((((((---(.((........)).)))))).)))).)))))))))))))))))))))))))))).... ( -35.10, z-score = -1.85, R) >droMoj3.scaffold_6680 18619756 119 + 24764193 UAGGUCGGGCUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUCGUCGAGCGAAAAGCGAAUUCAUUGUUGGAUUUUGUUGCUCUUGGUAUUCGAGCCAAUAAGUUCGGCAACCAGUU ..(((...((......((.(((((((.(((((.(((..(((((.....((((((.....(((((((....)))))))..))))))..)))))))))))))))))))))))).))).... ( -36.70, z-score = -1.95, R) >droGri2.scaffold_15110 14860069 100 - 24565398 UAUUUCCAGCUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUGGCUAAGGAAA-------------GUUGGAUUUC------CUAGGUAUUCGAGCCAAUAAGUUCGGCAGCGGGUU ....(((.(((....(((.(((((((.(((((.(((..((((((.....((((((-------------......))))------)))))))))))))))))))))))))).)))))).. ( -33.70, z-score = -2.75, R) >consensus UGGCCCGCACUCUUAUCGUAACUUGUGUGGCUUCGAAUGUACCUAGUUGAGGAAAAA_____UCCCAUGUCGGAUUUUGUUACCUCUGGUAUUCGAGCCAAUAAGUUCGGCCACGAAUU ...............(((.(((((((.(((((.(((..(((((.....((((.((.......((........)).....)).)))).)))))))))))))))))))))))......... (-23.29 = -23.95 + 0.66)


| Location | 11,766,096 – 11,766,210 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Shannon entropy | 0.31201 |
| G+C content | 0.45127 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.31 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11766096 114 - 24543557 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCGACAUGGGA-----GUUUUCCUCAACUAGGUAAGUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((.(((....(((......)))-----)))..)))).....)))..))))).)))))...))))).((.........))))))) ( -32.00, z-score = -1.47, R) >droSim1.chr3L 11131599 114 - 22553184 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCGACACGGGA-----CUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((....(((.(((......)))-----.))).)))).....))))..)))).)))))...))))).((.........))))))) ( -31.70, z-score = -1.78, R) >droSec1.super_0 3951119 114 - 21120651 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCGACACGGGA-----CUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((....(((.(((......)))-----.))).)))).....))))..)))).)))))...))))).((.........))))))) ( -31.70, z-score = -1.78, R) >droYak2.chr3L 11794901 114 - 24197627 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCGACAUGGGG-----UUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((....(((((((......)))-----)))).)))).....))))..)))).)))))...))))).((.........))))))) ( -34.80, z-score = -2.40, R) >droEre2.scaffold_4784 11743708 114 - 25762168 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCGACAUGGGA-----UUCUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((......(((((......)))-----))...)))).....))))..)))).)))))...))))).((.........))))))) ( -33.00, z-score = -2.10, R) >droAna3.scaffold_13337 6019502 112 - 23293914 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCAAACUG--A-----UUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ......(((((.(((((..(((((((((((((.((((....((((((......)--)-----)))).)))).....))))..)))).)))))...))))).((.........))))))) ( -31.30, z-score = -2.59, R) >dp4.chrXR_group8 4870686 113 + 9212921 AUUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCC-AUUUCGGA-----GUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGACCA ......(((((((((((..(((((((((((((.((((.(((....(((-.....)))-----)))..)))).....))))..)))).)))))...)))))((.......)).))).))) ( -29.60, z-score = -1.72, R) >droPer1.super_29 522786 113 + 1099123 AUUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCC-AUUUCGGA-----GUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGACCA ......(((((((((((..(((((((((((((.((((.(((....(((-.....)))-----)))..)))).....))))..)))).)))))...)))))((.......)).))).))) ( -29.60, z-score = -1.72, R) >droWil1.scaffold_180949 3805278 114 + 6375548 AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCAACAUUGAA-----UUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUACGAACUA ..((((((.((((((((..(((((((((((((.((((....(((((.((....)).)-----)))).)))).....))))..)))).)))))...))))).))....)..))))))... ( -25.60, z-score = -2.13, R) >droVir3.scaffold_13049 23988145 115 - 25233164 AACCGGUGGCCGAACUUAUUGGCUCGAAUACCAAAGGCAACAAAAUCCAAACGUUGAAU---UCGUUCCUCAACCAGGUACAUUC-AAGCCACACAAGUUACGAUAAGAGCACGAACCA ....(((.(((((((((..(((((.(((((((...(....)...........(((((..---.......)))))..))))..)))-.)))))...))))).))......))....))). ( -25.50, z-score = -1.50, R) >droMoj3.scaffold_6680 18619756 119 - 24764193 AACUGGUUGCCGAACUUAUUGGCUCGAAUACCAAGAGCAACAAAAUCCAACAAUGAAUUCGCUUUUCGCUCGACGAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGCCCGACCUA ....(((((.(((((((..(((((((((((((.(((((......................)))))(((.....)))))))..)))).)))))...))))).)).........))))).. ( -25.05, z-score = -0.96, R) >droGri2.scaffold_15110 14860069 100 + 24565398 AACCCGCUGCCGAACUUAUUGGCUCGAAUACCUAG------GAAAUCCAAC-------------UUUCCUUAGCCAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGCUGGAAAUA ..((.(((..(((((((..((((((((((((((((------((((......-------------)))))).....)))))..)))).)))))...))))).)).....))).))..... ( -28.80, z-score = -3.63, R) >consensus AAUUCGUGGCCGAACUUAUUGGCUCGAAUACCAGAGGUAACAAAAUCCAACAUGGGA_____UUUUUCCUCAACUAGGUACAUUCGAAGCCACACAAGUUACGAUAAGAGUGCGGGCCA ..........(((((((..(((((((((((((.((((..............................)))).....))))..)))).)))))...))))).))................ (-17.99 = -18.31 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:37 2011