
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,885,596 – 4,885,666 |
| Length | 70 |
| Max. P | 0.999762 |

| Location | 4,885,596 – 4,885,666 |
|---|---|
| Length | 70 |
| Sequences | 8 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 66.89 |
| Shannon entropy | 0.64242 |
| G+C content | 0.35145 |
| Mean single sequence MFE | -14.74 |
| Consensus MFE | -12.14 |
| Energy contribution | -11.56 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
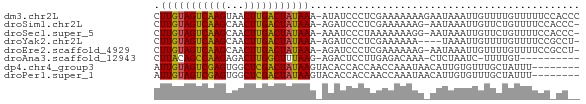
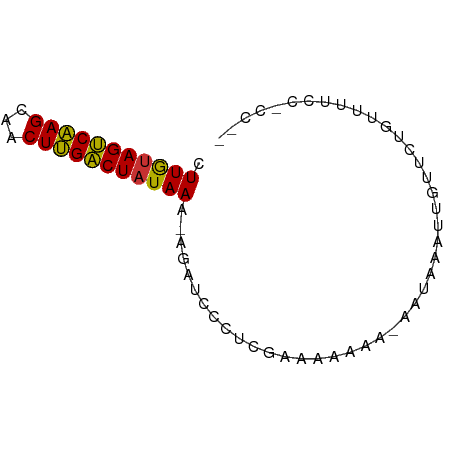
>dm3.chr2L 4885596 70 + 23011544 CUUGUAGUCAAGUAACUUGACUAUAAA-AUAUCCCUCGAAAAAAAGAAUAAAUUGUUUUGUUUUUCCACCC .(((((((((((...))))))))))).-.........((((((.(((((.....))))).))))))..... ( -15.60, z-score = -4.90, R) >droSim1.chr2L 4755643 68 + 22036055 CUUGUAGUCAAGCAACUUGACUAUAAA-AGAUCCCUCGAAAAAAG-AAUAAAUUGUUCUGUUUUCCACCC- .(((((((((((...))))))))))).-.........(((((.((-(((.....))))).))))).....- ( -17.30, z-score = -5.24, R) >droSec1.super_5 2980262 68 + 5866729 CUUGUAGUCAAGCAACUUGACUAUAAA-AAAUCCCUAAAAAAAGG-AAUAAAUUGUUCUGUUUUCCACCC- .(((((((((((...))))))))))).-...............((-((..............))))....- ( -13.84, z-score = -3.77, R) >droYak2.chr2L 4896318 65 + 22324452 CUUGUAGUCAAGCAACUUGACUAUAAA-AGAUCCCUCGAAAAAA----UAAAUUGUUUUGUUUUCCGCCU- .(((((((((((...))))))))))).-............((((----((((....))))))))......- ( -13.40, z-score = -3.79, R) >droEre2.scaffold_4929 4967430 68 + 26641161 CUUGUAGUCAAGCAACUUGACUAUAAA-AGAUCCCUCGAAAAAAG-AAUAAAUUGUUUUGUUUUCCGCCU- .(((((((((((...))))))))))).-.........(((((.((-(((.....))))).))))).....- ( -14.70, z-score = -3.76, R) >droAna3.scaffold_12943 2625479 58 + 5039921 CUUACAGCCAAGAGACUUGGCUUUAAG-AGACUCCUUGAGACAAA-CUCUAAUC-UUUUGU---------- ((((.(((((((...))))))).))))-.........(((.....-))).....-......---------- ( -13.10, z-score = -1.39, R) >dp4.chr4_group3 6134943 63 - 11692001 AUUGUAGUCGAGUGGCUCGACUAUAAGUACACCACCAACCAAAUAACAUUGUGUUUGCUAUUU-------- .(((((((((((...))))))))))).............((((((......))))))......-------- ( -15.00, z-score = -2.18, R) >droPer1.super_1 3233550 63 - 10282868 AUUGUAGUCGAGUGGCUCGACUAUAAGUACACCACCAACCAAAUAACAUUGUGUUUGCUAUUU-------- .(((((((((((...))))))))))).............((((((......))))))......-------- ( -15.00, z-score = -2.18, R) >consensus CUUGUAGUCAAGCAACUUGACUAUAAA_AGAUCCCUCGAAAAAAA_AAUAAAUUGUUCUGUUUUCC_CC__ .(((((((((((...)))))))))))............................................. (-12.14 = -11.56 + -0.58)
| Location | 4,885,596 – 4,885,666 |
|---|---|
| Length | 70 |
| Sequences | 8 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 66.89 |
| Shannon entropy | 0.64242 |
| G+C content | 0.35145 |
| Mean single sequence MFE | -13.24 |
| Consensus MFE | -8.62 |
| Energy contribution | -8.15 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
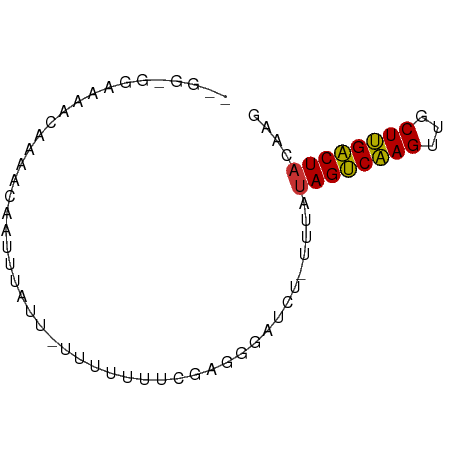
>dm3.chr2L 4885596 70 - 23011544 GGGUGGAAAAACAAAACAAUUUAUUCUUUUUUUCGAGGGAUAU-UUUAUAGUCAAGUUACUUGACUACAAG (..(....)..).....((..((((((((.....)))))))).-.)).((((((((...)))))))).... ( -12.30, z-score = -1.72, R) >droSim1.chr2L 4755643 68 - 22036055 -GGGUGGAAAACAGAACAAUUUAUU-CUUUUUUCGAGGGAUCU-UUUAUAGUCAAGUUGCUUGACUACAAG -.....(((((.((((.......))-)).))))).........-....((((((((...)))))))).... ( -12.40, z-score = -0.80, R) >droSec1.super_5 2980262 68 - 5866729 -GGGUGGAAAACAGAACAAUUUAUU-CCUUUUUUUAGGGAUUU-UUUAUAGUCAAGUUGCUUGACUACAAG -.....................(((-(((......))))))..-....((((((((...)))))))).... ( -12.20, z-score = -0.89, R) >droYak2.chr2L 4896318 65 - 22324452 -AGGCGGAAAACAAAACAAUUUA----UUUUUUCGAGGGAUCU-UUUAUAGUCAAGUUGCUUGACUACAAG -...(((((((............----.)))))))........-....((((((((...)))))))).... ( -10.32, z-score = -0.91, R) >droEre2.scaffold_4929 4967430 68 - 26641161 -AGGCGGAAAACAAAACAAUUUAUU-CUUUUUUCGAGGGAUCU-UUUAUAGUCAAGUUGCUUGACUACAAG -...(((((((..((........))-..)))))))........-....((((((((...)))))))).... ( -10.50, z-score = -0.58, R) >droAna3.scaffold_12943 2625479 58 - 5039921 ----------ACAAAA-GAUUAGAG-UUUGUCUCAAGGAGUCU-CUUAAAGCCAAGUCUCUUGGCUGUAAG ----------.....(-((((.(((-.....)))....)))))-((((.(((((((...))))))).)))) ( -14.80, z-score = -1.73, R) >dp4.chr4_group3 6134943 63 + 11692001 --------AAAUAGCAAACACAAUGUUAUUUGGUUGGUGGUGUACUUAUAGUCGAGCCACUCGACUACAAU --------((((((((.......))))))))((((((((((..((.....))...))))).)))))..... ( -16.70, z-score = -2.25, R) >droPer1.super_1 3233550 63 + 10282868 --------AAAUAGCAAACACAAUGUUAUUUGGUUGGUGGUGUACUUAUAGUCGAGCCACUCGACUACAAU --------((((((((.......))))))))((((((((((..((.....))...))))).)))))..... ( -16.70, z-score = -2.25, R) >consensus __GG_GGAAAACAAAACAAUUUAUU_UUUUUUUCGAGGGAUCU_UUUAUAGUCAAGUUGCUUGACUACAAG ................................................((((((((...)))))))).... ( -8.62 = -8.15 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:41 2011