| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,709,312 – 11,709,453 |
| Length | 141 |
| Max. P | 0.730954 |

| Location | 11,709,312 – 11,709,453 |
|---|---|
| Length | 141 |
| Sequences | 8 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.44711 |
| G+C content | 0.46714 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
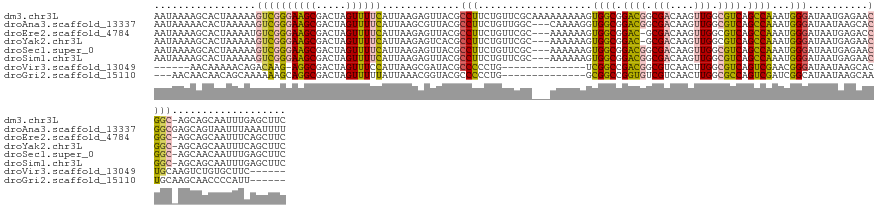
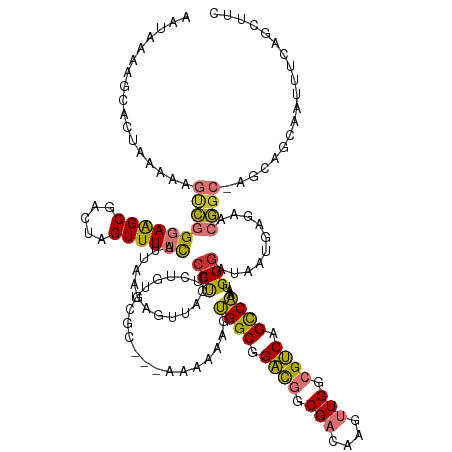
>dm3.chr3L 11709312 141 - 24543557 AAUAAAAGCACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGAGUUACGCCUUCUGUUCGCAAAAAAAAAGUGGCGGACGGCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGAACGGC-AGCAGCAAUUUGAGCUUC .....((((.......(((((.....))))).((((((((((.......((((.(((((.(((.........)))))))).)))).(...(((((....)))))...)..)))))))))).((-....)).......)))). ( -42.60, z-score = -2.40, R) >droAna3.scaffold_13337 5973179 139 - 23293914 AAUAAAAACACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGCGUUACGCCUUCUGUUGGC---CAAAAGGUGGCGGACGGCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUAAGCACGGCGAGCAGUAAUUUAAAUUUU .........(((.....((((((((((.....)))))).....((....((((.((((.(.((---(....))).))))).)))).(...(((((....)))))...)........)).))))....)))............ ( -37.50, z-score = -1.03, R) >droEre2.scaffold_4784 11692379 137 - 25762168 AAUAAAAGCACUAAAAUGUCGGGAAGCGACUAGUUUUCAUUAAGAGUUACGCCUUCUGUUCGC---AAAAAAGUGGCGGAC-GCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGACCGGC-AGCAGCAAUUUCAGCUUC .....((((.(((...(((((((((((.....)))))).......(((.((((..((......---.....)).)))))))-.)))))..(((((....))))).))).....(((((...((-....))..))))))))). ( -35.30, z-score = -0.17, R) >droYak2.chr3L 11742326 137 - 24197627 AAUAAAAGCACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGAGUCACGCCUUCUGUUCGC---AAAAAAGUGGCGGAC-GCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGAACGGC-AGCAGCAAUUUCAGCUUC .....((((.......(((((.....))))).((((((((((.(.(((.((((..((......---.....)).)))))))-.)..(...(((((....)))))...)..)))))))))).((-....)).......)))). ( -38.80, z-score = -1.46, R) >droSec1.super_0 3895759 138 - 21120651 AAUAAAAGCACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGAGUUACGCCUUCUGUUCGC---AAAAAAGUGGCGGACGGCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGAACGGC-AGCAACAAUUUGAGCUUC .....((((.......(((((.....))))).((((((((((.......((((.(((((.(((---......)))))))).)))).(...(((((....)))))...)..))))))))))...-.............)))). ( -43.00, z-score = -2.85, R) >droSim1.chr3L 11078894 138 - 22553184 AAUAAAAGCACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGAGUUACGCCUUCUGUUCGC---AAAAAAGUGGCGGACGGCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGAACGGC-AGCAGCAAUUUGAGCUUC .....((((.......(((((.....))))).((((((((((.......((((.(((((.(((---......)))))))).)))).(...(((((....)))))...)..)))))))))).((-....)).......)))). ( -43.60, z-score = -2.64, R) >droVir3.scaffold_13049 23925545 115 - 25233164 ------AACAAAAACAGACAAG-AGGCGACUAGUUUCCAUUAAGCGAUACGCCCCCUG--------------UCGGCCGACGGCGUCAACUUGGCGUCAGUCGAACGGGAUAAUAAGCACUGCAAGUCUGUGCUUC------ ------.......((((((...-.((((....((((.....))))....)))).((((--------------(((((.((((.((......)).)))).)))).)))))................)))))).....------ ( -37.30, z-score = -1.86, R) >droGri2.scaffold_15110 14808717 119 + 24565398 ---AACAACAACAGCAAAAAAGCAGGCGACUAGUUUUUAUUAAACGGUACGCCCCCUG--------------GCGGCCGGUGUCGUCAACUUGGCGCCAGUCGAUCGGCAUAAUAAGCAAUGCAAGCAACCCCAUU------ ---..................((.(((((((.((((.....))))(((.((((....)--------------))))))(((((((......)))))))))))).)).((((........))))..)).........------ ( -33.70, z-score = -0.85, R) >consensus AAUAAAAGCACUAAAAAGUCGGGAAGCGACUAGUUUUCAUUAAGAGUUACGCCUUCUGUUCGC___AAAAAAGUGGCGGACGGCGACAAGUUGGCGUCAGCCAAAUGGGAUAAUGAGAACGGC_AGCAGCAAUUUCAGCUUC .........................((.....((.(((((((.......((((.(((((................))))).)))).(...(((((....)))))...)..))))))).)).....))............... (-20.56 = -21.35 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:32 2011