
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,694,449 – 11,694,557 |
| Length | 108 |
| Max. P | 0.654805 |

| Location | 11,694,449 – 11,694,557 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.66080 |
| G+C content | 0.48920 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.14 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
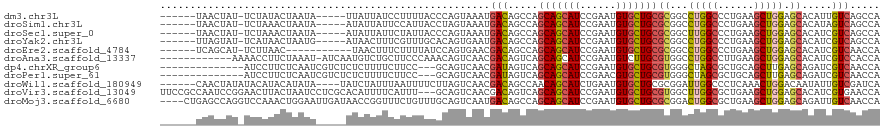
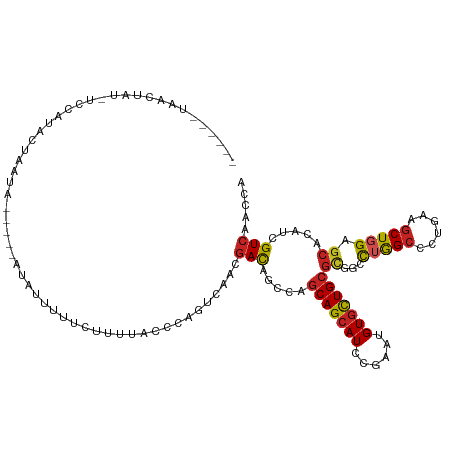
>dm3.chr3L 11694449 108 - 24543557 ------UAACUAU-UCUAUACUAAUA-----UUAUUAUCCUUUUACCCAGUAAAUGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCCUGGCCCUGAAGCUGGAGCACAUUGUCAGCCA ------.......-............-----.......................((((((((.(((((((......)))))))..)))(..((......))..).......))))).... ( -26.10, z-score = -0.46, R) >droSim1.chr3L 11064295 108 - 22553184 ------UAACUAU-UCUAAACUAAUA-----AUAUUAUUCCAUUACCUAGUAAAUGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCCUGGCCCUGAAGCUGGAGCACAUAGUCAGCCA ------.......-.....((((.((-----((........))))..))))...((((.(((.(((((((......)))))))..)))(..((......))..)........)))).... ( -26.30, z-score = -0.63, R) >droSec1.super_0 3880784 108 - 21120651 ------UAACUAU-UCUAAACUAAUA-----AUAUUAUUCUAUUACCCAGUAAAUGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCUUGGCCCUGAAGCUGGAGCACAUCGUCAGCCA ------.......-.....(((..((-----(((......)))))...)))...((((((((.(((((((......)))))))..))))((((((.......)).)).))..)))).... ( -26.70, z-score = -0.81, R) >droYak2.chr3L 11724967 108 - 24197627 ------UUAGUAU-UCAUAACUAAUG-----AUAACUUUCGUUUGCACAGUGAAUGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCCUGGCCCUGAAGCUGGAGCACAUCGUCAGCCA ------.....((-((((....((((-----(......)))))......))))))(((.(((.(((((((......)))))))..)))(..((......))..)........)))..... ( -30.40, z-score = -0.15, R) >droEre2.scaffold_4784 11677350 102 - 25762168 ------UCAGCAU-UCUUAAC-----------UAACUUUCUUUUAUCCAGUGAACGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCCUGGCCCUGAAGCUGGAGCACAUCGUCAACCA ------.......-.......-----------.............((((((......(((((.(((((((......)))))))..)).)))........))))))............... ( -26.24, z-score = -0.13, R) >droAna3.scaffold_13337 5959467 107 - 23293914 ------------AAAACCUUCUAAAU-AUCAAUGUCUGCUUCCCAAACAGUCAACGACAGUCAGCAGCAUCCGAAUGUCUUGCGUGGCCUGGCCUUGAAGCUGGAGCACAUCGUCCACCA ------------..............-.....((.(((.........))).))..(((.....(((((((....))))..)))((((((..((......))..).)).))).)))..... ( -18.30, z-score = 1.60, R) >dp4.chrXR_group6 3316346 103 + 13314419 --------------AUCCUUCUCAAUCGUCUCUCUUUUCUUCC---GCAGUCAACGAUAGUCAGCAGCAUCCGAAUGUGCUGCGUGGGCUAGCGCUGCAGCUUGAGCAGAUCGUCAACCA --------------...((.(((((.((.....).........---(((((......(((((.(((((((......)))))))...)))))..))))).).))))).))........... ( -27.00, z-score = 0.01, R) >droPer1.super_61 71712 103 + 350175 --------------AUCCUUCUCAAUCGUCUCUCUUUUCUUCC---GCAGUCAACGAUAGUCAGCAGCAUCCGAACGUGCUGCGUGGGCUAGCGCUGCAGCUUGAGCAGAUCGUCAACCA --------------...((.(((((.((.....).........---(((((......(((((.(((((((......)))))))...)))))..))))).).))))).))........... ( -27.10, z-score = -0.04, R) >droWil1.scaffold_180949 3722151 110 + 6375548 ------CAACUAUAUACAUACAUAUA----UAUCUAUUUAAUUUUCUUAGUCAACGACAGCCAACAGCAUCUGAAUGUGCUGCGCGGAUUGGCCCUCAAACUGGACAAUAUUGUCGAUCA ------....((((((......))))----))......................((((((((((((((((......))))))......)))))((.......)).......))))).... ( -19.90, z-score = -0.47, R) >droVir3.scaffold_13049 23912458 117 - 25233164 UUCCGCCAAUCCGGAACUUACUAAUCCUCGCACAUUUUCAUUU---GCAGUCAACGACAGUCAGCAGCAUCCGAAUGUGCUGCGUGGCUUGGCGCUGAAGCUGGAGCACAUCGUGAACCA (((((......)))))...........((((.((.(((((...---((.((((.....((((((((((((......))))))).)))))))))))))))).))((.....)))))).... ( -31.20, z-score = 0.37, R) >droMoj3.scaffold_6680 18537751 116 - 24764193 ----CUGAGCCAGGUCCAAACUGGAAUUGAUAACCGGUUUCUGUUUGCAGUCAAUGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGACUGGCGCUGAAGCUGGAGCAGAUUGUCAACCA ----(((..((((...(((((.(((((((.....))))))).)))))...(((.((.(((((.(((((((......)))))))..)).))).)).)))..))))..)))........... ( -40.20, z-score = -0.88, R) >consensus ______UAACUAU_UCCAUACUAAUA_____AUAUUUUUCUUUUACCCAGUCAACGACAGCCAGCAGCAUCCGAAUGUGCUGCGCGGCCUGGCCCUGAAGCUGGAGCACAUCGUCAACCA .......................................................(((.....(((((((......)))))))((...(((((......))))).)).....)))..... (-17.50 = -17.45 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:30 2011