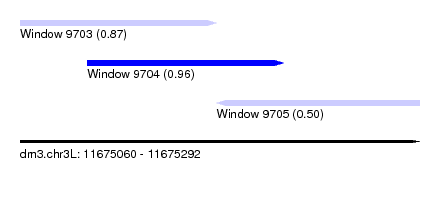
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,675,060 – 11,675,292 |
| Length | 232 |
| Max. P | 0.956651 |

| Location | 11,675,060 – 11,675,174 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.97 |
| Shannon entropy | 0.61055 |
| G+C content | 0.39975 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.51 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11675060 114 + 24543557 CAAGUGUUUCACCUAAAGGCGAGGCUUAUUUAGGGGUUUUCG-CGUUGCUUCGAGGACUUUCGAAGGAAUUUCUUUGGCCACAUUGCUGCACAAAGAAGAAUUUGGCGGUGGAAA ......(((((((((((((((((((((......))).)))))-)(((.(((((((....))))))).)))..)))))((((.(((.((......))...))).))))))))))). ( -33.30, z-score = -0.92, R) >droAna3.scaffold_13337 5940564 105 + 23293914 ----AAUUCCACAUGAAAAUUAAAAAAACGUA-CGCUUUCUAACACUACU----AGGCUGUGGUAUUAUUGUCAUCACUAACACCAAUAAAAACAUACGCGGU-GGCGGCGCAAA ----........................(((.-(((....(((.(((((.----.....))))).))).............((((...............)))-))))))).... ( -14.66, z-score = 0.86, R) >droEre2.scaffold_4784 11657497 114 + 25762168 GAAGUUUUCCACCGAAUGGUGUGGUAUAUUUAGGGUUUUUCUGCAUUGCUCUG-UAAUCUUCGCAGUAAUUACCUUGGCCACAUUGAUGCACAAAAAAGAAUUUGGCGGUGGAAA .....((((((((....((((((((.....((((((..(((((((((((...)-))))....)))).))..))))))))))))))...((.((((......)))))))))))))) ( -36.30, z-score = -2.94, R) >droYak2.chr3L 11703650 114 + 24197627 GAAGUUUUCCACCUACUGCUGAGGUUUAUUUAGUGCUUU-CUACAUUGCGUUGGUAAUUUUCGAAGCAAUUAACUUGGCCACAUUGAUGCACAAAAGGGAAUUUGGCGGUGGAAA .....((((((((.........((((...((((((((((-..(.(((((....))))).)..))))).)))))...))))........((.((((......)))))))))))))) ( -28.70, z-score = -0.69, R) >droSec1.super_0 3861255 114 + 21120651 AAAGUGUUUUACCCAAUGUCGAGGCUUAUUUAAGGUUUUUCU-CAUUGCUUCGACAAAAUUCGAAGUAAUAACUUUGGCCACAUUGCUGCACAAAAAAGAAUUUGGCGGUGGAAA ......(((((((((((((...((((.....((((((.....-.((((((((((......))))))))))))))))))))))))))..((.((((......))))))))))))). ( -30.60, z-score = -2.22, R) >droSim1.chr3L 11044790 114 + 22553184 AAAGUGUUUCACCCAAUGACGAGGCUUAUUUAGGGUUUUUCU-CAUUGCUUCGAGAAAUUUCGAAGUAAUAACUUUGGCCACAUUGCUGCACAAAAAAGAAUUUGGCGGUGGAAA ......((((((((((((....((((......(((.....))-)(((((((((((....)))))))))))......)))).)))))..((.((((......))))))))))))). ( -32.20, z-score = -2.22, R) >consensus AAAGUGUUCCACCUAAUGACGAGGCUUAUUUAGGGUUUUUCU_CAUUGCUUCGAGAAAUUUCGAAGUAAUUACUUUGGCCACAUUGAUGCACAAAAAAGAAUUUGGCGGUGGAAA ......(((((((...............................((((((((((......)))))))))).......((((.(((..............))).))))))))))). (-13.84 = -15.51 + 1.67)
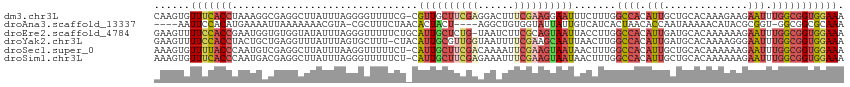
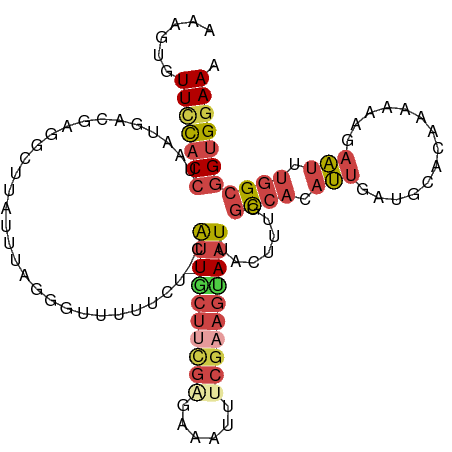
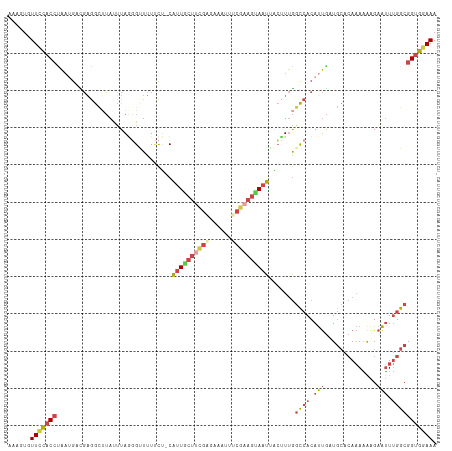
| Location | 11,675,099 – 11,675,213 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.69 |
| Shannon entropy | 0.55446 |
| G+C content | 0.38411 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11675099 114 + 24543557 UCGCGUUGCUUCGAGGACUUUCGAAGGAAUUUCUUUGGCCA-CAUUGCUGCACAAAGAAGAAUUUGGCGGUGGAAAUUUU-AUUUUUGCCGUGGAAAAUCAUACGAAAUACGGGCA ..(((((.(((((((....))))))).)))(((((((((..-.......)).)))))))...(((.((((..(((((...-)))))..)))).))).................)). ( -30.90, z-score = -1.01, R) >droAna3.scaffold_13337 5940599 108 + 23293914 -----UAACACUACUAGGCUGUGGUAUUAUUGUCAUCACUAACACCAAUAAAAACAUACGCGGU-GGCGGCGCAAAUUUUUAGUUUCUCCUUGGAAAAUUGGAAAAACUACUGC-- -----.........(((((((((((.(((.((....)).))).)))........(((.....))-)))))).........((((((.(((..........))).))))))))).-- ( -18.90, z-score = 0.13, R) >droEre2.scaffold_4784 11657537 113 + 25762168 CUGCAUUGCUCUG-UAAUCUUCGCAGUAAUUACCUUGGCCA-CAUUGAUGCACAAAAAAGAAUUUGGCGGUGGAAAUUUU-AUUUUUGCCGUUGAAAAGCGUACAAAAUACGGCCA (((((((((...)-))))....)))).........(((((.-..(((((((.(......)..((..((((..(((((...-)))))..))))..))..)))).))).....))))) ( -29.00, z-score = -1.18, R) >droYak2.chr3L 11703689 114 + 24197627 CUACAUUGCGUUGGUAAUUUUCGAAGCAAUUAACUUGGCCA-CAUUGAUGCACAAAAGGGAAUUUGGCGGUGGAAAUUUU-AUUUUUGCCGUGCAAAAACUUAACAAAUAGGGUCA ....(((((.((((......)))).))))).....(((((.-......(((.((((......))))((((..(((((...-)))))..)))))))....((........))))))) ( -26.00, z-score = -0.64, R) >droSec1.super_0 3861294 114 + 21120651 UCUCAUUGCUUCGACAAAAUUCGAAGUAAUAACUUUGGCCA-CAUUGCUGCACAAAAAAGAAUUUGGCGGUGGAAAUUUU-AUUUUAGUCGUGGAAAAUCAUAGGAAAUACGGUCA ....((((((((((......)))))))))).....(((((.-..(..((((.((((......))))))))..)......(-(((((....(((......)))..)))))).))))) ( -28.90, z-score = -2.60, R) >droSim1.chr3L 11044829 114 + 22553184 UCUCAUUGCUUCGAGAAAUUUCGAAGUAAUAACUUUGGCCA-CAUUGCUGCACAAAAAAGAAUUUGGCGGUGGAAAUUUU-AUUUUAGUCGUGGAAAAUCUUAUGAAAUACGGUCA ....(((((((((((....))))))))))).....(((((.-..(..((((.((((......))))))))..).......-.......((((((......)))))).....))))) ( -30.00, z-score = -2.70, R) >consensus UCGCAUUGCUUCGAUAAAUUUCGAAGUAAUUACCUUGGCCA_CAUUGAUGCACAAAAAAGAAUUUGGCGGUGGAAAUUUU_AUUUUUGCCGUGGAAAAUCAUAAGAAAUACGGUCA ....((((((((((......))))))))))......((((............((((......))))((((..(((((....)))))..))))...................)))). (-17.12 = -17.77 + 0.65)
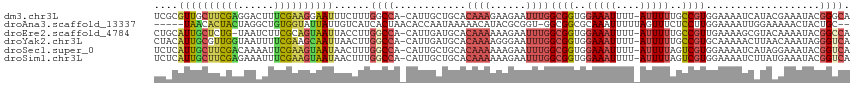

| Location | 11,675,174 – 11,675,292 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.81 |
| Shannon entropy | 0.43011 |
| G+C content | 0.44448 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -18.09 |
| Energy contribution | -20.23 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 11675174 118 - 24543557 -GAGCACUUUGCGGUUAUUCAAUGCAGGCAGUGGGUCCGUGGAUCUUCAUGGGUCUACCGGCAGUCAUCUGCUCGCUGUUUGCCCGUAUUUCGUAUGAUUUUCCACGGCAAAAAUAAAA- -......(((((.((........(((((((((((..((((((....))))))..)....(((((....))))))))))))))).(((((...))))).......)).))))).......- ( -37.30, z-score = -1.72, R) >droAna3.scaffold_13337 5940669 116 - 23293914 GCAACCCUUGGCCUUAAUUAAAUUUCUUUAG--AGCCUAUAUCACCUCAUAUGUUUUUUGUAGCUCAACUGCCUUCUAUU--GCAGUAGUUUUUCCAAUUUUCCAAGGAGAAACUAAAAA ((((.....(((......((((....))))(--(((.((((.................))))))))....))).....))--))..((((((((((..........)))))))))).... ( -23.13, z-score = -2.30, R) >droEre2.scaffold_4784 11657611 118 - 25762168 -GAGCACUCUUCGGUUAUUCAAUGCAGGCAGUGGGUCCGUGGGUCUUCAUGGGUCUACCGGCAAUCAUCUGCUCCCUGUUUGGCCGUAUUUUGUACGCUUUUCAACGGCAAAAAUAAAA- -(.(((.....((((((......(((((..((((..((((((....))))))..)))).((((......)))).))))).)))))).....))).)(((.......)))..........- ( -32.60, z-score = -0.85, R) >droYak2.chr3L 11703764 118 - 24197627 -GAGCACUUUUUGGUUAUUCAAUGCAGGCCGUGGGUCUGUGGAUCUUCAUGGGUCUACCGGCAGUCAUCUGCUCCCUGUUUGACCCUAUUUGUUAAGUUUUUGCACGGCAAAAAUAAAA- -..(((....((((....)))))))..((((((((((.((((((((....)))))))).(((((....)))))........)))))..........((....)))))))..........- ( -34.50, z-score = -1.36, R) >droSec1.super_0 3861369 118 - 21120651 -GAGCACUUUGCGGUUAUUCAAUGCAGGCAGUGGGUUCGUGGAUCUUCAUGCGUCUACCGGCAGUCAUCUGCUCCCUGUUUGACCGUAUUUCCUAUGAUUUUCCACGACUAAAAUAAAA- -........((((((((......(((((..(((((..(((((....)))))..))))).(((((....))))).))))).))))))))...............................- ( -29.30, z-score = -0.47, R) >droSim1.chr3L 11044904 118 - 22553184 -GAGCACUUUGCGGUUAUUCAAUGCAGGCAGUGGGUUCGUGGAUCUUCAUGGGUCUACCGGCAGUCAUCUGCUCCCUGUUUGACCGUAUUUCAUAAGAUUUUCCACGACUAAAAUAAAA- -........((((((((......(((((..(((((..(((((....)))))..))))).(((((....))))).))))).))))))))...............................- ( -32.90, z-score = -1.37, R) >consensus _GAGCACUUUGCGGUUAUUCAAUGCAGGCAGUGGGUCCGUGGAUCUUCAUGGGUCUACCGGCAGUCAUCUGCUCCCUGUUUGACCGUAUUUCGUAAGAUUUUCCACGGCAAAAAUAAAA_ .........((((((((......(((((..((((((((((((....)))))))))))).(((((....))))).))))).))))))))................................ (-18.09 = -20.23 + 2.15)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:29 2011