| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,667,867 – 11,667,958 |
| Length | 91 |
| Max. P | 0.867120 |

| Location | 11,667,867 – 11,667,958 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.66088 |
| G+C content | 0.68712 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -13.70 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
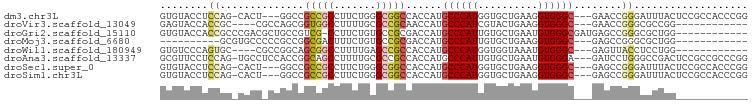
>dm3.chr3L 11667867 91 + 24543557 GUGUACCUCCAG-CACU---GGCCGCCGCCUUCUGGGCGGCCACCAUGCCCAUGGUGCUGAAGGUGGGC---GAACCGGGAUUUACUCCGCCACCCGG ((.(((((.(((-(...---(((((((........)))))))(((((....))))))))).))))).))---...(((((.............))))) ( -45.52, z-score = -2.68, R) >droVir3.scaffold_13049 23883354 79 + 25233164 GAGUACCACCGC----CGCCAGCGGUGGCCUUUUGCGCCGCAACCAUGCCCAUCGUACUGAAGGUGGGC---GAACCGGGCGCCGG------------ ........(((.----((((.((((((........)))))).....((((((((........)))))))---).....)))).)))------------ ( -36.30, z-score = -1.32, R) >droGri2.scaffold_15110 10777806 85 - 24565398 GUGUACCACCGCCCGACGCUGCCGUCG-CCUUCUGUGCCGCGACCAUGCCCAUUGUGCUGAAUGUGGGCGAUGAGCCGGGCGCUGG------------ .....(((.((((((..(((...((((-(..........)))))(((((((((..........)))))).)))))))))))).)))------------ ( -34.90, z-score = -1.25, R) >droMoj3.scaffold_6680 18509648 73 + 24764193 ----------GCGUGCCCCCGCCGGCGACUUUCUGUGCCGCGACCAUGCCCAUUGUGCUGAAGGUGGGC---GAGCCGGGCGCUGG------------ ----------..((((((.(((.(((.((.....))))))))....((((((((........)))))))---)....))))))...------------ ( -32.50, z-score = -0.50, R) >droWil1.scaffold_180949 3690715 79 - 6375548 GUGUCCCAGUGC----CGCCGGCAGCGGCCUUUUGAGCCGCCACCAUGCCCAUGGUGGUAAAUGUGGGC---GAGUUACCUCCUGG------------ .....((((.((----((((.(((((((((....).)))))((((((....)))))).....))).)))---).))......))))------------ ( -33.20, z-score = -1.48, R) >droAna3.scaffold_13337 5933876 94 + 23293914 GCGUUCCUCCAG-UGCCUCCACCGGCAGCCUUUUGCGCCGCCACCAUGCCCAUUGUGCUGAAUGUGGGA---GAUCCUGGGCCGACUCCGCCGCCCGG ......((((..-((((......))))((.(((.((((.((......)).....)))).))).)).)))---)...((((((((....))..)))))) ( -26.00, z-score = 1.80, R) >droSec1.super_0 3854459 91 + 21120651 GUGUACCUCCAG-CACU---GGCCGCCGCCUUCUGGGCGGCCACCAUGCCCAUGGUGCUGAAGGUGGGC---GAGCCGGGAUUUACUCCGCCACCCGG ((.(((((.(((-(...---(((((((........)))))))(((((....))))))))).))))).))---...(((((.............))))) ( -45.22, z-score = -2.08, R) >droSim1.chr3L 11037968 91 + 22553184 GUGUACCUCCAG-CACU---GGCCGCCGCCUUCUGGGCGGCCACCAUGCCCAUGGUGCUGAAGGUGGGC---GAGCCGGGAUUUACUCCGCCACCCGG ((.(((((.(((-(...---(((((((........)))))))(((((....))))))))).))))).))---...(((((.............))))) ( -45.22, z-score = -2.08, R) >consensus GUGUACCUCCAC_CACC_CCGGCGGCGGCCUUCUGGGCCGCCACCAUGCCCAUGGUGCUGAAGGUGGGC___GAGCCGGGAGCUAC____________ ........((..............(((((.......)))))......((((((..........))))))........))................... (-13.70 = -13.88 + 0.17)

| Location | 11,667,867 – 11,667,958 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.66088 |
| G+C content | 0.68712 |
| Mean single sequence MFE | -41.02 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
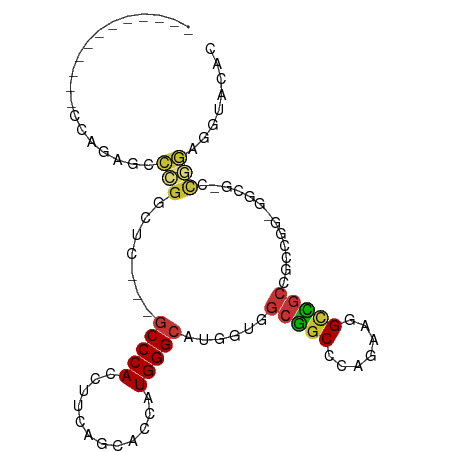
>dm3.chr3L 11667867 91 - 24543557 CCGGGUGGCGGAGUAAAUCCCGGUUC---GCCCACCUUCAGCACCAUGGGCAUGGUGGCCGCCCAGAAGGCGGCGGCC---AGUG-CUGGAGGUACAC ..(((((((((........)))..))---))))(((((((((((........((((.((((((.....)))))).)))---))))-)))))))).... ( -50.00, z-score = -3.18, R) >droVir3.scaffold_13049 23883354 79 - 25233164 ------------CCGGCGCCCGGUUC---GCCCACCUUCAGUACGAUGGGCAUGGUUGCGGCGCAAAAGGCCACCGCUGGCG----GCGGUGGUACUC ------------...((((((..((.---(((((.(........).)))))..))..).))))).....((((((((.....----)))))))).... ( -35.60, z-score = -0.39, R) >droGri2.scaffold_15110 10777806 85 + 24565398 ------------CCAGCGCCCGGCUCAUCGCCCACAUUCAGCACAAUGGGCAUGGUCGCGGCACAGAAGG-CGACGGCAGCGUCGGGCGGUGGUACAC ------------(((.((((((((.(((.(((((............))))))))((((((.(......).-)).))))...)))))))).)))..... ( -39.40, z-score = -2.65, R) >droMoj3.scaffold_6680 18509648 73 - 24764193 ------------CCAGCGCCCGGCUC---GCCCACCUUCAGCACAAUGGGCAUGGUCGCGGCACAGAAAGUCGCCGGCGGGGGCACGC---------- ------------...((.((((.(..---(((((............)))))......(((((.......))))).).)))).))....---------- ( -29.00, z-score = 0.29, R) >droWil1.scaffold_180949 3690715 79 + 6375548 ------------CCAGGAGGUAACUC---GCCCACAUUUACCACCAUGGGCAUGGUGGCGGCUCAAAAGGCCGCUGCCGGCG----GCACUGGGACAC ------------(((((((....)))---(((........((.....))((.(((..((((((.....))))))..))))))----)).))))..... ( -37.20, z-score = -2.78, R) >droAna3.scaffold_13337 5933876 94 - 23293914 CCGGGCGGCGGAGUCGGCCCAGGAUC---UCCCACAUUCAGCACAAUGGGCAUGGUGGCGGCGCAAAAGGCUGCCGGUGGAGGCA-CUGGAGGAACGC ((((.((((...)))).))(((..((---(((.((..(((((.......)).)))(((((((.......))))))))))))))..-)))..))..... ( -34.00, z-score = 0.99, R) >droSec1.super_0 3854459 91 - 21120651 CCGGGUGGCGGAGUAAAUCCCGGCUC---GCCCACCUUCAGCACCAUGGGCAUGGUGGCCGCCCAGAAGGCGGCGGCC---AGUG-CUGGAGGUACAC ..((((((((((.....)))..)).)---))))(((((((((((........((((.((((((.....)))))).)))---))))-)))))))).... ( -51.50, z-score = -3.27, R) >droSim1.chr3L 11037968 91 - 22553184 CCGGGUGGCGGAGUAAAUCCCGGCUC---GCCCACCUUCAGCACCAUGGGCAUGGUGGCCGCCCAGAAGGCGGCGGCC---AGUG-CUGGAGGUACAC ..((((((((((.....)))..)).)---))))(((((((((((........((((.((((((.....)))))).)))---))))-)))))))).... ( -51.50, z-score = -3.27, R) >consensus ____________CCAGAGCCCGGCUC___GCCCACCUUCAGCACCAUGGGCAUGGUGGCGGCCCAGAAGGCCGCCGCCGG_GGCG_CCGGAGGUACAC ...................(((.......(((((............)))))......(((((.......))))).............)))........ (-14.41 = -14.25 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:23 2011