| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,654,660 – 11,654,752 |
| Length | 92 |
| Max. P | 0.760701 |

| Location | 11,654,660 – 11,654,752 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.92 |
| Shannon entropy | 0.56233 |
| G+C content | 0.56135 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
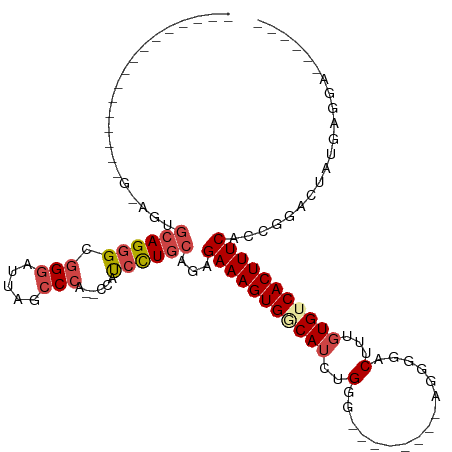
>dm3.chr3L 11654660 92 - 24543557 --------------AGUCAGCGCAGGGCGGGAUUACCCCACCCCAUCCUGCAGAGAAAGUGGCAUCUGGG--------AGGCGACUUUGUGUCACUUUCACCCGACUAUGAGGA------ --------------((((...(((((..(((..........)))..)))))...(((((((((((...((--------((....)))))))))))))))....)))).......------ ( -37.40, z-score = -2.23, R) >droSim1.chr3L 11024217 83 - 22553184 ---------------------GCAGGGCGGGAUUAGCCCA--CCAUCCUGCAGAGAAAGUGGCAUCUGGG--------AGGGGACUUUGUGUCACUUUCACCGGACUGUGAGGA------ ---------------------((((.(((((((.......--..)))))))...(((((((((((...((--------((....)))))))))))))))......)))).....------ ( -32.70, z-score = -1.44, R) >droSec1.super_0 3841433 83 - 21120651 ---------------------GCAGGGCGGGAUUAGCCCA--CCAUCCUGCAGAGAAAGUGGCAUCUGGG--------AGGGGACUUUGUGUCACUUUCACCGGACUAUGAGGA------ ---------------------((((((.(((.....))).--...))))))...(((((((((((...((--------((....))))))))))))))).((.........)).------ ( -32.60, z-score = -1.85, R) >droYak2.chr3L 11683207 102 - 24197627 --------------GAGUAGUGCAGGGCGGGAUUAGCCCA--CCAUCCUGCAGAGAAAGUGGCAUCUGUUCCCCAGUCCACAGACUUUGUG-CACUUUCACCGGACUACAGGAAUGGGG- --------------..(((((.(.(((((((((.......--..)))))))...(((((((((((((((..........)))))...))).-))))))).))).)))))..........- ( -31.90, z-score = 0.08, R) >droEre2.scaffold_4784 11636704 88 - 25762168 --------------AGGCAGUGCAGGGCGGGAUUACCCCA--ACAUCCUGCGGAGAAAGUGGCAUCUGUG---------UGUGACUUGGUG-CACUUUCACCGGACUUUGGGGC------ --------------..((...((((((.(((.....))).--...))))))((.(((((((.((((.((.---------....))..))))-))))))).))..........))------ ( -29.30, z-score = 0.26, R) >droAna3.scaffold_13337 5922049 110 - 23293914 GCCCACGUGCAGCCUAGUAUUAUAGAGUAGUUCCCAUCCACAUACCCUUCCAGGGAAAGUGACAUCUG----------CAGGGACUUUCUGUCACUUUCGCCAAAUAGACAGGAUGGGGG .(((..(.((.(((((......))).)).)).)((((((......((.....))(((((((((((((.----------...))).....))))))))))............))))))))) ( -31.70, z-score = -0.62, R) >consensus ________________G_AGUGCAGGGCGGGAUUAGCCCA__CCAUCCUGCAGAGAAAGUGGCAUCUGGG________AGGGGACUUUGUGUCACUUUCACCGGACUAUGAGGA______ ........................(((((((((...........)))))))...(((((((((((.......................))))))))))).)).................. (-18.46 = -19.43 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:20 2011