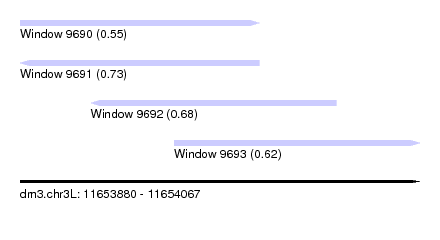
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,653,880 – 11,654,067 |
| Length | 187 |
| Max. P | 0.734732 |

| Location | 11,653,880 – 11,653,992 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.43660 |
| G+C content | 0.52331 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
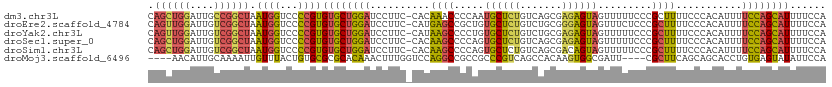
>dm3.chr3L 11653880 112 + 24543557 CAGCUGGAUUGCCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC-CACAAACCCCAAUGCUCUGUCAGCGAGAGUAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA ..((((((...(((((..((((...))))..))))).......-..........((((.......((((.(((......))).)))).......)))).))))))........ ( -27.64, z-score = -1.17, R) >droEre2.scaffold_4784 11635879 112 + 25762168 CAGUUGGAUUGUCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC-CAUGAGCCGCUGUGCUCUGUCUGCGGGAGUAGUUUCUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA ..((((((..(.(((((.((((...(((.....)))......)-))).))))))((((........(((((((......)))))))......))))...))))))........ ( -37.94, z-score = -2.87, R) >droYak2.chr3L 11682391 112 + 24197627 CAGUUGGAUUGUCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC-CAUAAGCCCCUGUGCUCUGUCUGCGAGAGUAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA ..((((((.(((.((((.((((...(((.....)))......)-))).))))...(((......((((....)))).......))).......)))...))))))........ ( -27.32, z-score = -1.24, R) >droSec1.super_0 3840679 112 + 21120651 CAGCUGGAUUGUCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC-CACAAGCCCCAGUGCUCUGUCAGCGAGAGUAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA ..((((((.(((.(((..((((...))))(..((((...(((.-...)))..))))..)...)))((((.(((......))).))))......)))...))))))........ ( -30.60, z-score = -1.78, R) >droSim1.chr3L 11023472 112 + 22553184 CAGCUGGAUUGUCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC-CACAAGCCCCAGUGCUCUGUCAGCGACAGUAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA ..((((((.(((.((..(((((.......(..((((...(((.-...)))..))))..).(((((...))))).........))).))..)).)))...))))))........ ( -30.00, z-score = -1.88, R) >droMoj3.scaffold_6496 18811875 105 + 26866924 ----AACAUUGCAAAAUUGUUUACUGUGCGCGCACAAACUUUGGUCCAGGCCGCCGCCCGUCAGCCACAAGUGGCGAUU----CGCUUCAGCAGCACCUGUGAGUAUAUUCCA ----...............(((((.((((..((.........(((....)))...((..(((.((((....))))))).----.))....)).))))..)))))......... ( -25.30, z-score = 1.01, R) >consensus CAGCUGGAUUGUCGGCUAAUGGUCCCCGUGUGCUGGAUCCUUC_CACAAGCCCCAGUGCUCUGUCAGCGAGAGUAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCA .((((((....)))))).((((...))))((((((((..........((((.....((((((.......)))))).........))))...........))))))))...... (-17.41 = -17.17 + -0.24)

| Location | 11,653,880 – 11,653,992 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.43660 |
| G+C content | 0.52331 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
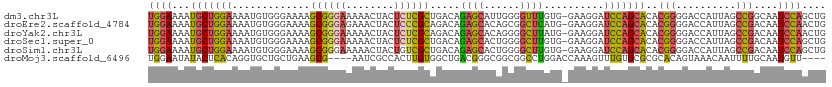
>dm3.chr3L 11653880 112 - 24543557 UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCUGACAGAGCAUUGGGGUUUGUG-GAAGGAUCCAGCACACGGGGACCAUUAGCCGGCAAUCCAGCUG ((((...((((((..(((((......(((((((........)))))))......)))))((..((((((-(..(....)..).))))))..)).....)))))).)))).... ( -35.70, z-score = -1.65, R) >droEre2.scaffold_4784 11635879 112 - 25762168 UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAGAAACUACUCCCGCAGACAGAGCACAGCGGCUCAUG-GAAGGAUCCAGCACACGGGGACCAUUAGCCGACAAUCCAACUG ((((...(((((((.............(((((((......)))))))...((((((......)))).))-......)))))))..(((..........)))....)))).... ( -38.60, z-score = -3.19, R) >droYak2.chr3L 11682391 112 - 24197627 UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCAGACAGAGCACAGGGGCUUAUG-GAAGGAUCCAGCACACGGGGACCAUUAGCCGACAAUCCAACUG ((((...(((((((.............((((((........))))))...((((((......)))).))-......)))))))..(((..........)))....)))).... ( -30.10, z-score = -1.33, R) >droSec1.super_0 3840679 112 - 21120651 UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCUGACAGAGCACUGGGGCUUGUG-GAAGGAUCCAGCACACGGGGACCAUUAGCCGACAAUCCAGCUG ........((((((...(((.((...(((((((........)))))))..........(((..((((((-(..(....)..).))))))..)))....)).))).)))))).. ( -36.20, z-score = -2.03, R) >droSim1.chr3L 11023472 112 - 22553184 UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUGUCGCUGACAGAGCACUGGGGCUUGUG-GAAGGAUCCAGCACACGGGGACCAUUAGCCGACAAUCCAGCUG ........((((((...(((.((....((((......)).(((((...))))).))..(((..((((((-(..(....)..).))))))..)))....)).))).)))))).. ( -33.80, z-score = -1.30, R) >droMoj3.scaffold_6496 18811875 105 - 26866924 UGGAAUAUACUCACAGGUGCUGCUGAAGCG----AAUCGCCACUUGUGGCUGACGGGCGGCGGCCUGGACCAAAGUUUGUGCGCGCACAGUAAACAAUUUUGCAAUGUU---- (((.......((.(((((((((((((....----..))((((....)))).....)))))).)))))))))).....((((....))))(((((....)))))......---- ( -29.21, z-score = 1.20, R) >consensus UGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCUGACAGAGCACUGGGGCUUGUG_GAAGGAUCCAGCACACGGGGACCAUUAGCCGACAAUCCAGCUG ((((...(((((((.............((((((........)))))).....((((......))))..........)))))))..(((..........)))....)))).... (-20.96 = -21.22 + 0.26)
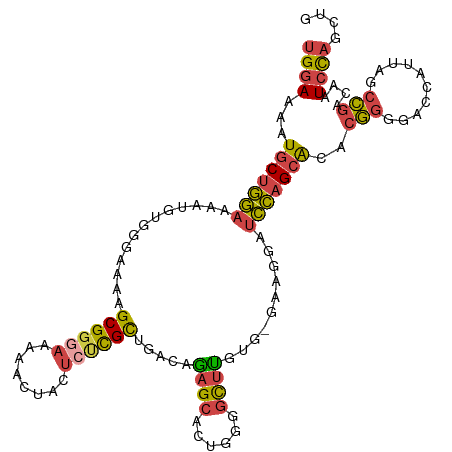
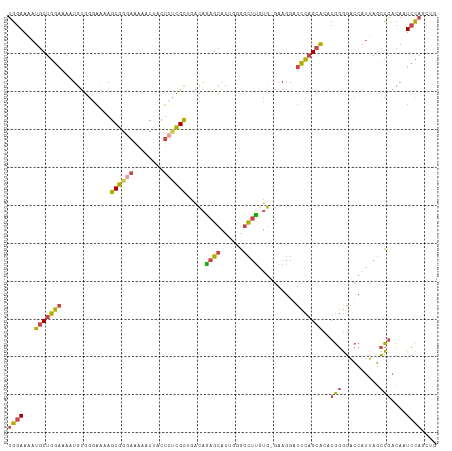
| Location | 11,653,913 – 11,654,028 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.40 |
| Shannon entropy | 0.50364 |
| G+C content | 0.53186 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11653913 115 - 24543557 CACACACACACGGUGGUCUGCGCCACAUGUUGUGCGUGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCUGACAGAGCAUUGGGGUUUGUGGAAGGAUCCA ..((((.((.(((((.(((((.((((((.....((((.....))))......))))))....(((((((........)))))))).)))).)))))..)).)))).......... ( -39.60, z-score = -2.14, R) >droEre2.scaffold_4784 11635912 113 - 25762168 --CACACACACGGUGGUCCGCGCCGAAUGUUGUGCGUGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAGAAACUACUCCCGCAGACAGAGCACAGCGGCUCAUGGAAGGAUCCA --((((....(((((.((((((((((...))).)))))))...))))).....))))(((...(((((((......)))))))...((((((......)))).))......))). ( -44.40, z-score = -2.96, R) >droYak2.chr3L 11682424 109 - 24197627 ------CACACGGUGGUCCGCGGCAAAUGUAGUGCGUGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCAGACAGAGCACAGGGGCUUAUGGAAGGAUCCA ------(((((((((.((((((.((.......))))))))...))))).....))))(((...((((((........))))))...((((((......)))).))......))). ( -30.60, z-score = -0.79, R) >droSim1.chr3L 11023505 109 - 22553184 ------CACACGGUGGUCUGCGCCAAAUGUUGUGCGUGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUGUCGCUGACAGAGCACUGGGGCUUGUGGAAGGAUCCA ------....(((((.(((((.(((....((..((((.....))))..)).....)))....(((((.(........).)))))).)))).)))))((((((.....))).))). ( -29.40, z-score = 0.14, R) >droMoj3.scaffold_6496 18811905 105 - 26866924 ------UGGACUUUGUACUUCAUCCGACUUUGUGGGUGGAAUAUACUCACAGGUGCUGCUGAAGCG----AAUCGCCACUUGUGGCUGACGGGCGGCGGCCUGGACCAAAGUUUG ------.((((((((...((((((((......))))))))......((.(((((((((((((....----..))((((....)))).....)))))).))))))).)))))))). ( -37.70, z-score = -1.88, R) >consensus ______CACACGGUGGUCUGCGCCAAAUGUUGUGCGUGGAAAAUGCUGGAAAAUGUGGGAAAAGCGGGAAAAACUACUCUCGCAGACAGAGCACAGGGGCUUGUGGAAGGAUCCA ......(((((((((.(((((((..........)))))))...))))).....))))(((...((((((........)))))).....((((......)))).........))). (-17.42 = -18.90 + 1.48)

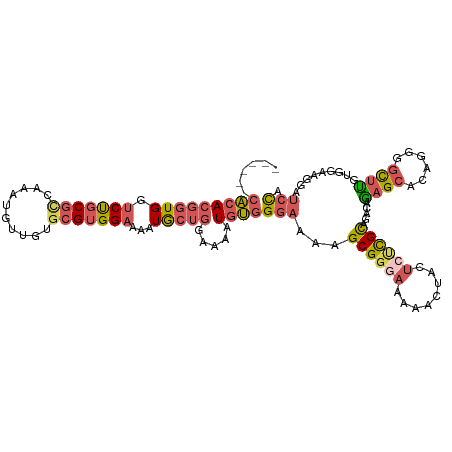
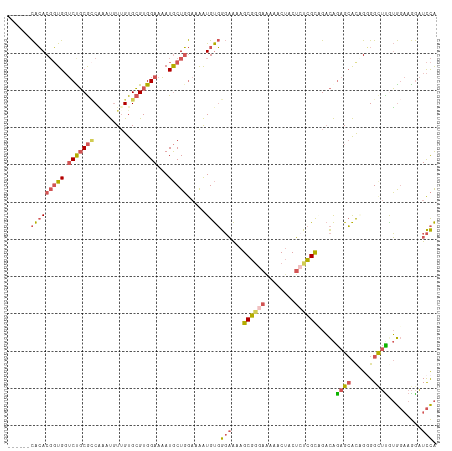
| Location | 11,653,952 – 11,654,067 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Shannon entropy | 0.42758 |
| G+C content | 0.48876 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -16.56 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11653952 115 + 24543557 UAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCACGCACAACAUGUGGCGCAGACCACCGUGUGUGUGUGUGUGUGUGAAUUUGAUUGGCAGUGAGCCGUAGAGCUUCU .((((((....((((...(((((..(((..((((....((((((((.(((((((......))).)))))))))))).))))..)))..))..)))....))))...))))))... ( -33.30, z-score = -1.48, R) >droEre2.scaffold_4784 11635951 113 + 25762168 UAGUUUCUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCACGCACAACAUUCGGCGCGGACCACCGUGUGUGUGUGUGUGUG--AAUUUGAUUGGCAGUGAGCCGGAGAGCUCCU .((((.((((.(((.....(((.....((((((..(((((((((((.(((...((((((....))))))))))))))))).)--))..)).))))..)))))).))))))))... ( -41.50, z-score = -3.42, R) >droYak2.chr3L 11682463 109 + 24197627 UAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCACGCACUACAUUUGCCGCGGACCACCGUGUGUGUGUGUG------AAUUUGAUUGGCAGUGAGCCGUGGAGCUUCU ...........(((....((((.........((..((((((((((.........(((((....))))).))))))).)------))..))...(((.....)))))))))).... ( -30.60, z-score = -1.72, R) >droSim1.chr3L 11023544 109 + 22553184 UAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCACGCACAACAUUUGGCGCAGACCACCGUGUGUGUGUGUG------AAUUUGAUUGGCAGUGAGCCGUAGAGCUUCU ...........(((((...(.((((..((((((..(((((((((((.(((.(((......)))..))))))))))).)------))..)).)))).)))).)....))))).... ( -29.80, z-score = -1.94, R) >droMoj3.scaffold_6496 18811942 106 + 26866924 UGGCGAUU--CGCUUCAGCAGCACCUGUGAGUAUAUUCCACCCACAAAGUCGGAUGAAGUACAAAGUCCACGUUGCCA-----GAACCUGUUAAUUAG--AACUAUGGAGCUUCU ........--.(((((((((((...((((.((.......)).))))..((.((((..........)))))))))))..-----....(((.....)))--.....)))))).... ( -21.80, z-score = 0.41, R) >consensus UAGUUUUUCCCGCUUUUCCCACAUUUUCCAGCAUUUUCCACGCACAACAUUUGGCGCAGACCACCGUGUGUGUGUGUG______AAUUUGAUUGGCAGUGAGCCGUAGAGCUUCU ...........(((((......................(((((((.((((.(((.(.....).))).)))))))))))...............(((.....)))..))))).... (-16.56 = -17.16 + 0.60)

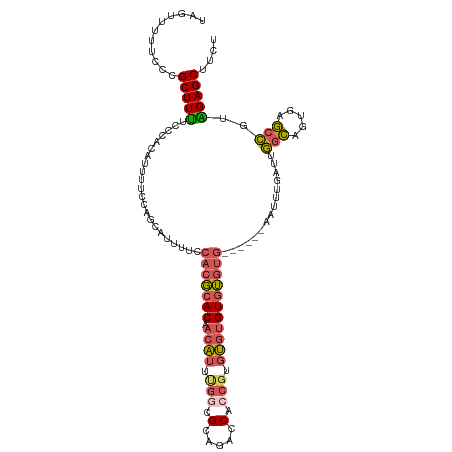
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:19 2011