| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,650,895 – 11,650,992 |
| Length | 97 |
| Max. P | 0.972018 |

| Location | 11,650,895 – 11,650,992 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.93 |
| Shannon entropy | 0.41829 |
| G+C content | 0.47562 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
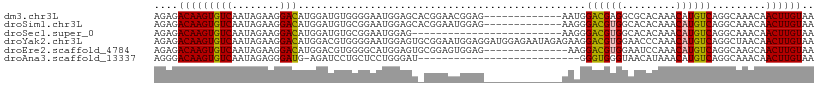
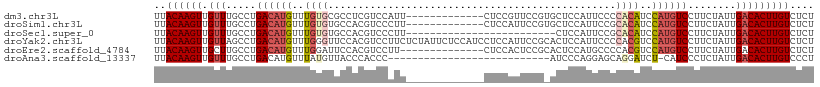
>dm3.chr3L 11650895 97 + 24543557 AGAGACAAGUGUCAAUAGAAGGACAUGGAUGUGGGGAAUGGAGCACGGAACGGAG-------------AAUGGACGAGGCGCACAAACAUGUCAGGCAAACAACUUGUAA ....((((((...........((((((..((((.(...((.....))...((...-------------......))...).))))..)))))).........)))))).. ( -20.95, z-score = -1.83, R) >droSim1.chr3L 11020612 97 + 22553184 AGAGACAAGUGUCAAUAGAAGGACAUGGAUGUGCGGAAUGGAGCACGGAAUGGAG-------------AAGGGACGUGGCACACAAACAUGUCAGGCAAACAACUUGUAA ....(((((((((..........(((...(((((........)))))..)))...-------------....((((((.........)))))).))).....)))))).. ( -23.30, z-score = -2.23, R) >droSec1.super_0 3837861 85 + 21120651 AGAGACAAGUGUCAAUAGAAGGACAUGGAUGUGCGGAAUGGAG-------------------------AAGGGACGUGGCACACAAACAUGUCAGGCAAACAACUUGUAA ....((((((...........((((((..((((.(..(((...-------------------------......)))..).))))..)))))).........)))))).. ( -22.05, z-score = -2.76, R) >droYak2.chr3L 11679495 110 + 24197627 AGAGACAAGUGUCAAUAGAAGGACAUGGACGUGGGGAAUGGAGUGCGGAAUGGAGGAUGGAGAAUAGAGAAGGACGUGGAACCCAAACAUGUCAGGCUAACAACUUGUAA ....((((((.....(((...((((((....((((.......((.(.........................).))......))))..))))))...)))...)))))).. ( -24.03, z-score = -1.74, R) >droEre2.scaffold_4784 11633184 96 + 25762168 AGAGACAAGUGUCAAUAGAAGGACAUGGACGUGGGGCAUGGAGUGCGGAGUGGAG--------------AAGGACGUGGAAUCCAAACAUGUCAGGCAAGCAACUUGUAA ....((((((...........((((((....((((.((((.....(........)--------------.....))))...))))..)))))).........)))))).. ( -21.55, z-score = -0.62, R) >droAna3.scaffold_13337 5919423 82 + 23293914 AGGGACAAGUGUCAAUAGAGGGAUG-AGAUCCUGCUCCUGGGAU---------------------------GGGUGGGUAACAUAAACAUGUCAGGCAAACAACUUGUAA ....((((((......((.(((((.-..))))).))(((((.((---------------------------(.(((.....)))...))).)))))......)))))).. ( -21.70, z-score = -0.81, R) >consensus AGAGACAAGUGUCAAUAGAAGGACAUGGAUGUGGGGAAUGGAGU_CGGAAUGGAG_____________AAAGGACGUGGAACACAAACAUGUCAGGCAAACAACUUGUAA ....(((((((((........)))................................................((((((.........)))))).........)))))).. (-12.77 = -12.68 + -0.08)
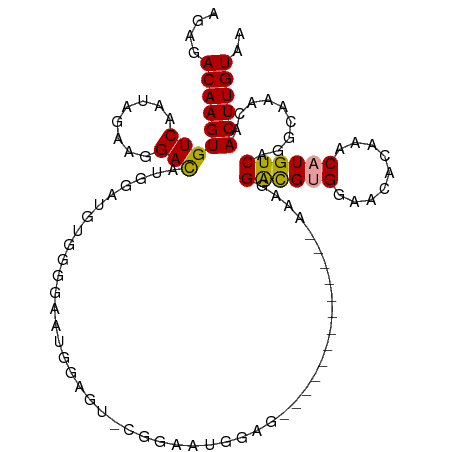
| Location | 11,650,895 – 11,650,992 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.93 |
| Shannon entropy | 0.41829 |
| G+C content | 0.47562 |
| Mean single sequence MFE | -17.63 |
| Consensus MFE | -10.14 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11650895 97 - 24543557 UUACAAGUUGUUUGCCUGACAUGUUUGUGCGCCUCGUCCAUU-------------CUCCGUUCCGUGCUCCAUUCCCCACAUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((.(((.....((((((..(((((((..((......-------------...))....))))..........)))..))))))........))))))))).... ( -17.82, z-score = -2.90, R) >droSim1.chr3L 11020612 97 - 22553184 UUACAAGUUGUUUGCCUGACAUGUUUGUGUGCCACGUCCCUU-------------CUCCAUUCCGUGCUCCAUUCCGCACAUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((.(((.....((((((..((((((.((((......-------------........))))........))))))..))))))........))))))))).... ( -22.66, z-score = -4.16, R) >droSec1.super_0 3837861 85 - 21120651 UUACAAGUUGUUUGCCUGACAUGUUUGUGUGCCACGUCCCUU-------------------------CUCCAUUCCGCACAUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((.(((.....((((((..((((((...........-------------------------........))))))..))))))........))))))))).... ( -20.73, z-score = -4.14, R) >droYak2.chr3L 11679495 110 - 24197627 UUACAAGUUGUUAGCCUGACAUGUUUGGGUUCCACGUCCUUCUCUAUUCUCCAUCCUCCAUUCCGCACUCCAUUCCCCACGUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((.(((((...((((((..((((..............................................))))....))))))......))))))))))).... ( -15.85, z-score = -1.57, R) >droEre2.scaffold_4784 11633184 96 - 25762168 UUACAAGUUGCUUGCCUGACAUGUUUGGAUUCCACGUCCUU--------------CUCCACUCCGCACUCCAUGCCCCACGUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((..(......((((((...((((.....))))..--------------.........(((.....)))........))))))........)..)))))).... ( -14.54, z-score = -0.81, R) >droAna3.scaffold_13337 5919423 82 - 23293914 UUACAAGUUGUUUGCCUGACAUGUUUAUGUUACCCACCC---------------------------AUCCCAGGAGCAGGAUCU-CAUCCCUCUAUUGACACUUGUCCCU ..((((((.(((((..((((((....))))))..))...---------------------------......((((..((((..-.))))))))...))))))))).... ( -14.20, z-score = -0.34, R) >consensus UUACAAGUUGUUUGCCUGACAUGUUUGUGUGCCACGUCCCUU_____________CUCCAUUCCG_ACUCCAUUCCCCACAUCCAUGUCCUUCUAUUGACACUUGUCUCU ..((((((.(((.....((((((..((((................................................))))..))))))........))))))))).... (-10.14 = -11.20 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:15 2011