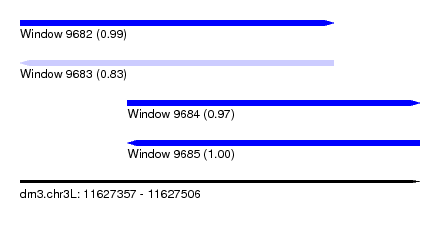
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,627,357 – 11,627,506 |
| Length | 149 |
| Max. P | 0.997247 |

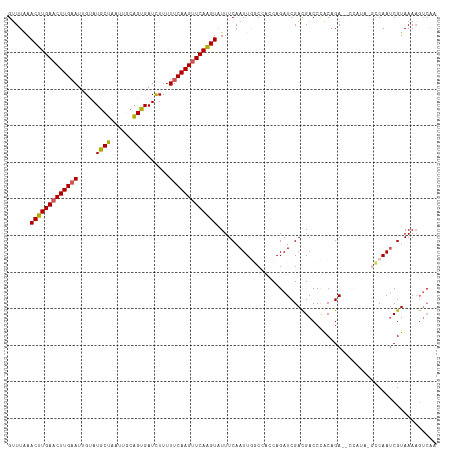
| Location | 11,627,357 – 11,627,474 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Shannon entropy | 0.28287 |
| G+C content | 0.37486 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -20.63 |
| Energy contribution | -20.51 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
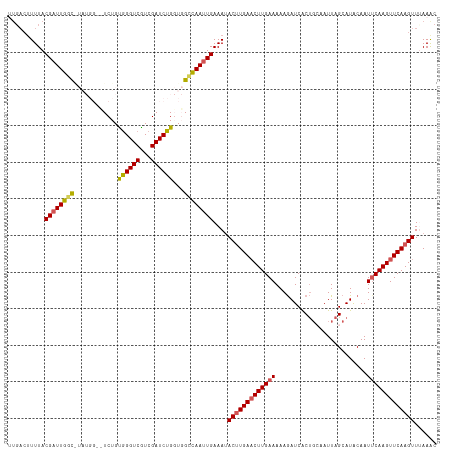
>dm3.chr3L 11627357 117 + 24543557 UUGACUUUUACGUUUGGCGUAUGGAUUCUGUGGGUCGUCGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAAAUCACCGCAAUUAGCAUACAAUUCAAGUUCAAGUUCAAAC ((((((..((((.....)))).)).(((....((((..(.....)..))))....)))..(((((((((((((..........((.....))......))))))))))))))))).. ( -27.59, z-score = -1.17, R) >droSim1.chr3L 10999868 109 + 22553184 UUGACUUUUACGAUUGGC--------UCUGCGGGUCGUCGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUGCAAUUCAAGUUCAAGUUUAAAC ..........((((((((--------...(((((((...)))))))..))))))))....(((((((((((((......((.(((.....)))))...)))))))))))))...... ( -36.30, z-score = -4.83, R) >droSec1.super_0 3814606 103 + 21120651 ------UUUACGAUUGGC--------UCUGCGGGUCGUCGAUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAAC ------....((((((((--------...(((((((...)))))))..))))))))....(((((((((((((.........(((.....))).....)))))))))))))...... ( -35.24, z-score = -5.53, R) >droYak2.chr3L 11655882 116 + 24197627 CUGACUUUUACGAUUGGUAUAUGG-CUCUGUGGGUCGUCGAUCUGGUGGGUAAUUGAAAAACUUGAACUUGAAAAAGAUCGCUUGUAUUAGCAUACAAUUCAAGUUCAAGUUAAAAC ((.(((.....(((((....((((-((.....))))))))))).))).)).........((((((((((((((....((.(((......)))))....))))))))))))))..... ( -30.80, z-score = -2.96, R) >droEre2.scaffold_4784 11611873 115 + 25762168 UUGACUUUUACGAUUGCUAUAUGG-CUCUGUAGGUCAUCGAUCUGGUGGGUAAUUGAAAUACCUGAUCUUGAAAAAGAUCGCUGGUAUUAGCAUACAAU-CAAGUUCAAGUUUAAGC ..(((((..(((((((...(((((-(((..((((((...))))))..))))......(((((((((((((....)))))))..))))))..))))))))-)..))..)))))..... ( -29.50, z-score = -1.89, R) >consensus UUGACUUUUACGAUUGGC_UAUGG__UCUGUGGGUCGUCGAUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAAC ..........((((((((............((((((...))))))...))))))))....(((((((((((((.........................)))))))))))))...... (-20.63 = -20.51 + -0.12)

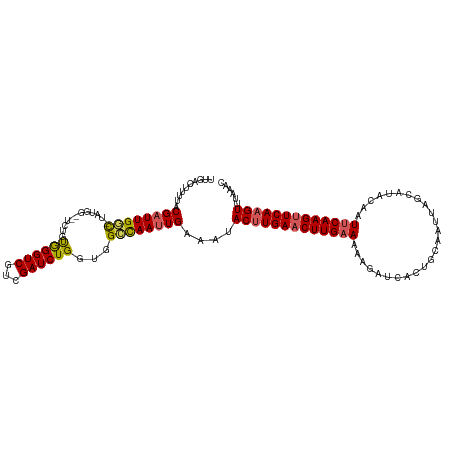
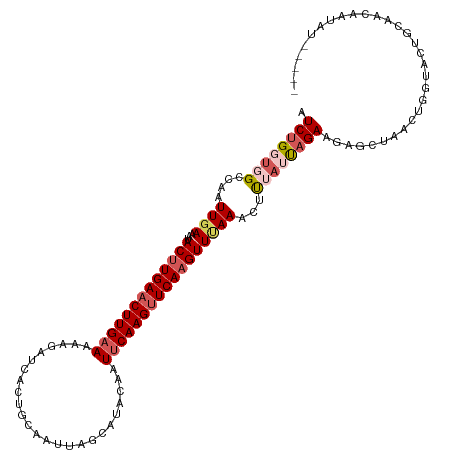
| Location | 11,627,357 – 11,627,474 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Shannon entropy | 0.28287 |
| G+C content | 0.37486 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11627357 117 - 24543557 GUUUGAACUUGAACUUGAAUUGUAUGCUAAUUGCGGUGAUUUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCGACGACCCACAGAAUCCAUACGCCAAACGUAAAAGUCAA ..(((((((((((((((((..((.(((.....)))...))....)))))))))))))...))))(((((.....((((...........).))).....)))))............. ( -25.90, z-score = -1.55, R) >droSim1.chr3L 10999868 109 - 22553184 GUUUAAACUUGAACUUGAAUUGCAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCGACGACCCGCAGA--------GCCAAUCGUAAAAGUCAA ......((((((((((((((..(.(((.....))))..)).....))))))))))))......((((((.......((..((...))..))--------))))))............ ( -26.11, z-score = -1.88, R) >droSec1.super_0 3814606 103 - 21120651 GUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAUCGACGACCCGCAGA--------GCCAAUCGUAAA------ ......(((((((((((((..(..(((.....))).....)...)))))))))))))......((((((.......((..((...))..))--------))))))......------ ( -24.51, z-score = -1.98, R) >droYak2.chr3L 11655882 116 - 24197627 GUUUUAACUUGAACUUGAAUUGUAUGCUAAUACAAGCGAUCUUUUUCAAGUUCAAGUUUUUCAAUUACCCACCAGAUCGACGACCCACAGAG-CCAUAUACCAAUCGUAAAAGUCAG .....((((((((((((((..(..((((......))))..)...))))))))))))))................(((..((((.........-...........))))....))).. ( -22.45, z-score = -3.20, R) >droEre2.scaffold_4784 11611873 115 - 25762168 GCUUAAACUUGAACUUG-AUUGUAUGCUAAUACCAGCGAUCUUUUUCAAGAUCAGGUAUUUCAAUUACCCACCAGAUCGAUGACCUACAGAG-CCAUAUAGCAAUCGUAAAAGUCAA ......((((.....((-(((((..(((......)))((((((....))))))((((...((.(((........))).))..))))......-.......)))))))...))))... ( -20.40, z-score = -1.18, R) >consensus GUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAUCGACGACCCACAGA__CCAUA_GCCAAUCGUAAAAGUCAA ......(((((((((((((.....((((......))))......)))))))))))))......((((((.......((...........))........))))))............ (-17.02 = -17.82 + 0.80)

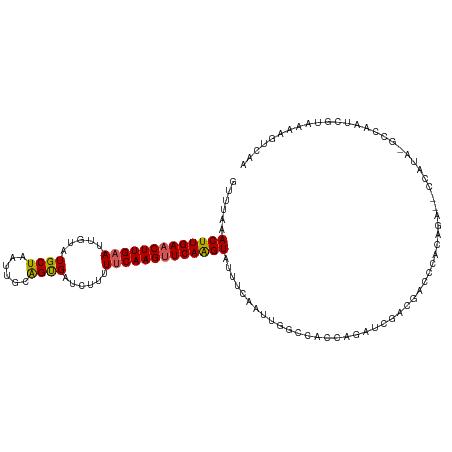
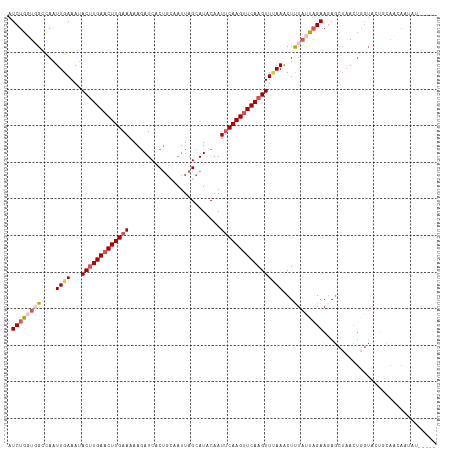
| Location | 11,627,397 – 11,627,506 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.39469 |
| G+C content | 0.31068 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -14.11 |
| Energy contribution | -15.51 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11627397 109 + 24543557 AUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAAAUCACCGCAAUUAGCAUACAAUUCAAGUUCAAGUUCAAACUCUAUUAGAAGAGCUAACUGGUAAAAAAUUAA-------- .((((((((....((((...(((((((((((((..........((.....))......)))))))))))))))))...)))))))).......................-------- ( -23.69, z-score = -2.85, R) >droSim1.chr3L 10999900 114 + 22553184 AUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUGCAAUUCAAGUUCAAGUUUAAACUUUACCAGAAGAGCUAACUGGUACUGCAACAAUAUAU--- ...((((.((..........(((((((((((((......((.(((.....)))))...)))))))))))))........((((((.........))))))..))))))......--- ( -26.60, z-score = -2.58, R) >droSec1.super_0 3814632 114 + 21120651 AUCUGUUUGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACUCUAUUUGAAGAGCUAACUGGUACUGCAACAAUAUAU--- ...(((.(((((.(((....(((((((((((((.........(((.....))).....))))))))))))).....((((......)))).))).)))))..))).........--- ( -25.84, z-score = -2.75, R) >droYak2.chr3L 11655921 95 + 24197627 AUCUGGUGGGUAAUUGAAAAACUUGAACUUGAAAAAGAUCGCUUGUAUUAGCAUACAAUUCAAGUUCAAGUUAAAACUUUCUUAGAAUAUUAUAU---------------------- .(((((..(((..((....((((((((((((((....((.(((......)))))....)))))))))))))).)))))..).)))).........---------------------- ( -21.90, z-score = -3.22, R) >droEre2.scaffold_4784 11611912 116 + 25762168 AUCUGGUGGGUAAUUGAAAUACCUGAUCUUGAAAAAGAUCGCUGGUAUUAGCAUACAAU-CAAGUUCAAGUUUAAGCUUUAUCAGAAGAGCUAAUUGGUACUGCAUGAAUAUUCUAU .......(((((.....(((((((((((((....)))))))..)))))).((((((...-..(((((((((....))))........))))).....))).))).....)))))... ( -24.50, z-score = -0.62, R) >consensus AUCUGGUGGCCAAUUGAAAUACUUGAACUUGAAAAAGAUCACUGCAAUUAGCAUACAAUUCAAGUUCAAGUUUAAACUUUAUUAGAAGAGCUAACUGGUACUGCAACAAUAU_____ .((((((((....((((...(((((((((((((.........................)))))))))))))))))...))))))))............................... (-14.11 = -15.51 + 1.40)

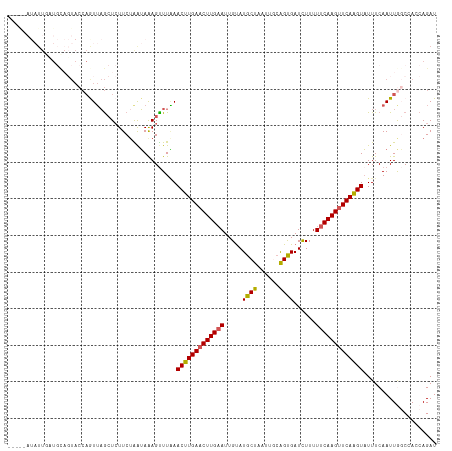
| Location | 11,627,397 – 11,627,506 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.39469 |
| G+C content | 0.31068 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11627397 109 - 24543557 --------UUAAUUUUUUACCAGUUAGCUCUUCUAAUAGAGUUUGAACUUGAACUUGAAUUGUAUGCUAAUUGCGGUGAUUUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAU --------...........((((((((((((......))))))...(((((((((((((..((.(((.....)))...))....))))))))))))).....))))))......... ( -29.40, z-score = -3.71, R) >droSim1.chr3L 10999900 114 - 22553184 ---AUAUAUUGUUGCAGUACCAGUUAGCUCUUCUGGUAAAGUUUAAACUUGAACUUGAAUUGCAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAU ---.....(((((((..((((((.........))))))........((((((((((((((..(.(((.....))))..)).....))))))))))))..........)).))))).. ( -32.80, z-score = -3.37, R) >droSec1.super_0 3814632 114 - 21120651 ---AUAUAUUGUUGCAGUACCAGUUAGCUCUUCAAAUAGAGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCAAACAGAU ---.....(((((......((((((((((((......))))))...(((((((((((((..(..(((.....))).....)...))))))))))))).....))))))..))))).. ( -31.00, z-score = -3.28, R) >droYak2.chr3L 11655921 95 - 24197627 ----------------------AUAUAAUAUUCUAAGAAAGUUUUAACUUGAACUUGAAUUGUAUGCUAAUACAAGCGAUCUUUUUCAAGUUCAAGUUUUUCAAUUACCCACCAGAU ----------------------..............((((.....((((((((((((((..(..((((......))))..)...))))))))))))))))))............... ( -21.50, z-score = -4.25, R) >droEre2.scaffold_4784 11611912 116 - 25762168 AUAGAAUAUUCAUGCAGUACCAAUUAGCUCUUCUGAUAAAGCUUAAACUUGAACUUG-AUUGUAUGCUAAUACCAGCGAUCUUUUUCAAGAUCAGGUAUUUCAAUUACCCACCAGAU ............((.((((((....((((..........))))....((((((...(-(((((............))))))...))))))....)))))).)).............. ( -19.30, z-score = -0.70, R) >consensus _____AUAUUGAUGCAGUACCAGUUAGCUCUUCUAAUAAAGUUUAAACUUGAACUUGAAUUGUAUGCUAAUUGCAGUGAUCUUUUUCAAGUUCAAGUAUUUCAAUUGGCCACCAGAU ..............................................(((((((((((((.....((((......))))......))))))))))))).................... (-16.88 = -16.72 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:12 2011