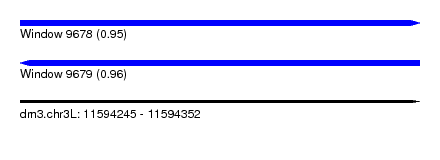
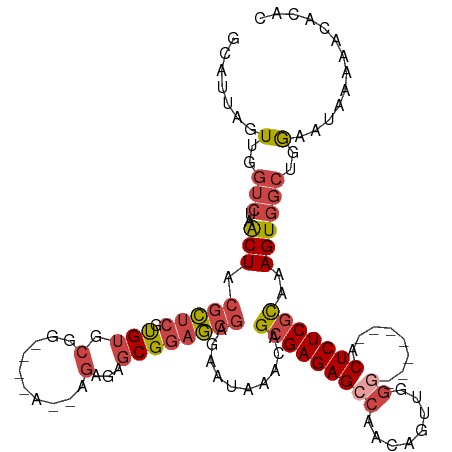
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,594,245 – 11,594,352 |
| Length | 107 |
| Max. P | 0.957906 |

| Location | 11,594,245 – 11,594,352 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.45285 |
| G+C content | 0.50684 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.79 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11594245 107 + 24543557 GUGUGUUUUUAUUCCAGCCACUUUGCGAGAGU-------UCCUACUGUUGGCUCUCGCUGUUUAUUCUCGCUCCGCUCUCU--U----CCGCACACGAGCGUAGUUAGACCAACUAAUGC (((.(((........))))))...((((((((-------(.........))))))))).(((...(((.(((.(((((...--.----........))))).))).)))..)))...... ( -27.10, z-score = -1.53, R) >droYak2.chr3L 11625601 114 + 24197627 GUGUGUUUUUAUGCCAGCCACUUUGCGAGAGUGUGGAGCCCCAAUUGUUGGCUCUCGCCGUUUAUUCUCGCUCCGCUCUCU--C----CCGCACGCGAGCGUAGUUAGACCAACUAAUGC ((((((..........((......))(((((.(((((((...(((...((((....))))...)))...))))))).))))--)----..))))))..((((((((.....)))).)))) ( -38.90, z-score = -3.00, R) >droEre2.scaffold_4784 11581201 111 + 25762168 GUGUGUUUUCAUUCCAGCCACUUUGCGAGAGC-------CCCAAUUGUUGGCUCUCGCUGUUUAUUCUCGCUCCGCUCUCU--CUCUGCCGCACACGAGCGUAGUUAGACCAACUAAUGC (((.(((........))))))...((((((((-------(.........))))))))).(((...(((.(((.(((((...--.............))))).))).)))..)))...... ( -31.29, z-score = -2.27, R) >droSec1.super_0 3783459 107 + 21120651 GUGUGUUUUUAUUCCAGCCACUUUGCGAGAGU-------CCCAACUGUUGGCUCUCGCUGUUUAUUCUCGCUCCGCUCUCU--U----CCGCACACGAGCGUAGUUAGACCAACUAAUGC (((.(((........))))))...((((((((-------(.........))))))))).(((...(((.(((.(((((...--.----........))))).))).)))..)))...... ( -28.90, z-score = -2.17, R) >droSim1.chr3L 10969948 107 + 22553184 GUCUGUUUUUAUUCCAGCCACUUUGCGAGAGU-------CCCAACUGUUGGCUCUCGCUGUUUAUUCUCGCUCCGCUCUCU--U----CCGCACACGAGCGUAGUUAGACCAACUAAUGC (((((...................((((((((-------(.........)))))))))...........(((.(((((...--.----........))))).)))))))).......... ( -27.00, z-score = -1.86, R) >droMoj3.scaffold_6680 18419350 100 + 24764193 UCAAGCGGUAAUUUUCACAACUUUAAGAGAGC-------------AACUAGCUCUCGCUAACUCUCUCCCACUCGCUGUCUGGCCUAUUCUCUCAUGCUCCUAGCAUGCACAA------- (((..((((.............(((.((((((-------------.....))))))..))).............))))..)))..........(((((.....))))).....------- ( -20.27, z-score = -1.34, R) >consensus GUGUGUUUUUAUUCCAGCCACUUUGCGAGAGU_______CCCAACUGUUGGCUCUCGCUGUUUAUUCUCGCUCCGCUCUCU__C____CCGCACACGAGCGUAGUUAGACCAACUAAUGC ........................((((((((..................)))))))).......(((.(((.(((((..................))))).))).)))........... (-16.32 = -17.79 + 1.47)

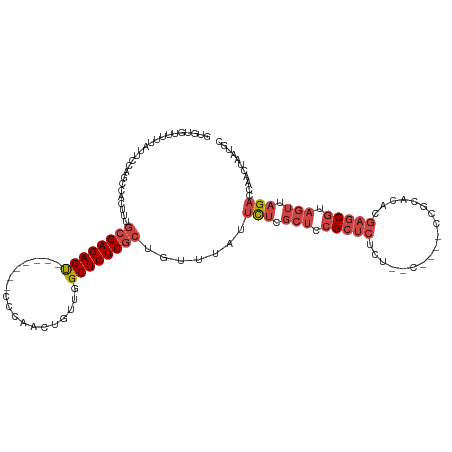
| Location | 11,594,245 – 11,594,352 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.45285 |
| G+C content | 0.50684 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -18.27 |
| Energy contribution | -20.11 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11594245 107 - 24543557 GCAUUAGUUGGUCUAACUACGCUCGUGUGCGG----A--AGAGAGCGGAGCGAGAAUAAACAGCGAGAGCCAACAGUAGGA-------ACUCUCGCAAAGUGGCUGGAAUAAAAACACAC ......(((..((((.((((((((.(((.(..----.--.)...)))))))...........(((((((((.......)).-------.)))))))...)))).)))).....))).... ( -32.40, z-score = -2.10, R) >droYak2.chr3L 11625601 114 - 24197627 GCAUUAGUUGGUCUAACUACGCUCGCGUGCGG----G--AGAGAGCGGAGCGAGAAUAAACGGCGAGAGCCAACAAUUGGGGCUCCACACUCUCGCAAAGUGGCUGGCAUAAAAACACAC ......((..(((..((((((....)))((((----(--((...(.(((((..........(((....)))..(....)..))))).).)))))))..))))))..))............ ( -37.50, z-score = -1.75, R) >droEre2.scaffold_4784 11581201 111 - 25762168 GCAUUAGUUGGUCUAACUACGCUCGUGUGCGGCAGAG--AGAGAGCGGAGCGAGAAUAAACAGCGAGAGCCAACAAUUGGG-------GCUCUCGCAAAGUGGCUGGAAUGAAAACACAC ......(((..((((.((((.(((((...(.((....--.....)).).)))))........(((((((((.........)-------))))))))...)))).)))).....))).... ( -36.90, z-score = -2.23, R) >droSec1.super_0 3783459 107 - 21120651 GCAUUAGUUGGUCUAACUACGCUCGUGUGCGG----A--AGAGAGCGGAGCGAGAAUAAACAGCGAGAGCCAACAGUUGGG-------ACUCUCGCAAAGUGGCUGGAAUAAAAACACAC ......(((..((((.((((((((.(((.(..----.--.)...)))))))...........(((((((((........))-------.)))))))...)))).)))).....))).... ( -35.00, z-score = -2.54, R) >droSim1.chr3L 10969948 107 - 22553184 GCAUUAGUUGGUCUAACUACGCUCGUGUGCGG----A--AGAGAGCGGAGCGAGAAUAAACAGCGAGAGCCAACAGUUGGG-------ACUCUCGCAAAGUGGCUGGAAUAAAAACAGAC ......(((..((((.((((((((.(((.(..----.--.)...)))))))...........(((((((((........))-------.)))))))...)))).)))).....))).... ( -35.00, z-score = -2.53, R) >droMoj3.scaffold_6680 18419350 100 - 24764193 -------UUGUGCAUGCUAGGAGCAUGAGAGAAUAGGCCAGACAGCGAGUGGGAGAGAGUUAGCGAGAGCUAGUU-------------GCUCUCUUAAAGUUGUGAAAAUUACCGCUUGA -------.....(((((.....)))))..................(((((((((((((((((((....))))...-------------))))))))..((((.....)))).))))))). ( -32.50, z-score = -2.37, R) >consensus GCAUUAGUUGGUCUAACUACGCUCGUGUGCGG____A__AGAGAGCGGAGCGAGAAUAAACAGCGAGAGCCAACAGUUGGG_______ACUCUCGCAAAGUGGCUGGAAUAAAAACACAC ......(((..((((.((((.(((((.(((..............)))..)))))........(((((((....................)))))))...)))).)))).....))).... (-18.27 = -20.11 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:07 2011