| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,591,330 – 11,591,422 |
| Length | 92 |
| Max. P | 0.975417 |

| Location | 11,591,330 – 11,591,422 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 67.73 |
| Shannon entropy | 0.59166 |
| G+C content | 0.62511 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796124 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
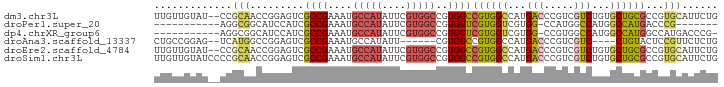
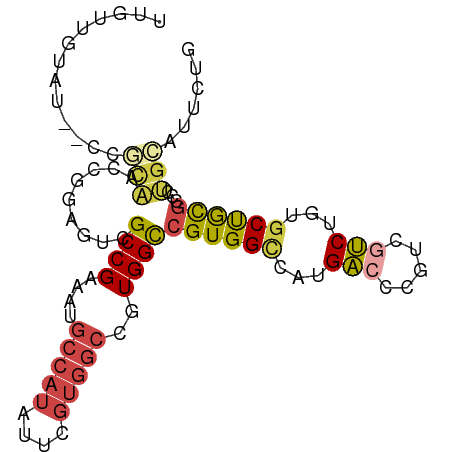
>dm3.chr3L 11591330 92 + 24543557 CAGAAUGCACGGCGCAGCACAGACGACGGGUCAUGGCCACGGCCACGGCCACGAAUAUGGCAUUUCGGCGACUCCGGUUGCGG--AUACAACAA .....(((.....)))((.((.....((((((.((((....)))).)))).))....))))...((.(((((....))))).)--)........ ( -29.20, z-score = -0.58, R) >droPer1.super_20 1839413 75 - 1872136 -------CGGGUCAUGGCCAUGGCCAUGG-CCACGACCACGACCACGGCCACGAAUAUGGCAUUUCGGCGAUGGAUGCCGCCU----------- -------((((((.(((((((....))))-))).)))).))......((((......)))).....((((........)))).----------- ( -32.40, z-score = -1.77, R) >dp4.chrXR_group6 3213801 81 - 13314419 -CGGGUCAUGGCCAUGGCCAUGGCCACGG-CCACGACCACGACCACGGCCACGAAUAUGGCAUUUCGGCGAUGGAUGCCGCCU----------- -((((((.(((((.((((....)))).))-))).)))).))......((((......)))).....((((........)))).----------- ( -38.60, z-score = -2.62, R) >droAna3.scaffold_13337 5867004 82 + 23293914 CAGAGAACGGAGUACAG----GACGACGGGUCAUGGCCACGGCCACG------AAUAUGGCAUUUCGGCGACUCCGGCCAUGA--CUCCGGCAG .......((((((.(((----(....((((((.((((....))))((------((........))))..))).))).)).)))--))))).... ( -32.00, z-score = -2.70, R) >droEre2.scaffold_4784 11578157 92 + 25762168 CAGAAUGCACGGCGCAGCACAGACGACGGGUCAUGGCCACGGCCACGGCCACGAAUAUGGCAUUUCGGCGACUCCGGUUGCGG--AUACAACAA .....(((.....)))((.((.....((((((.((((....)))).)))).))....))))...((.(((((....))))).)--)........ ( -29.20, z-score = -0.58, R) >droSim1.chr3L 10967275 94 + 22553184 CAGAAUGCACGGCGCAGCACAGACGACGGGUCAUGGCCACGGCCACGGCCACGAAUAUGGCAUUUCGGCGACUCCGGUUGCGGGGAUACAACAA .....(((.....)))((.((.....((((((.((((....)))).)))).))....))))((..(.(((((....))))).)..))....... ( -31.60, z-score = -1.05, R) >consensus CAGAAUGCACGGCACAGCACAGACGACGGGUCAUGGCCACGGCCACGGCCACGAAUAUGGCAUUUCGGCGACUCCGGCCGCGG__AUACAACAA ............((((((...(((.....)))...(((.((....))((((......)))).....))).......))))))............ (-13.48 = -14.53 + 1.06)

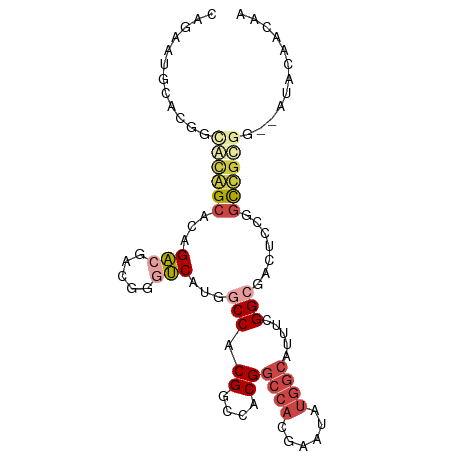
| Location | 11,591,330 – 11,591,422 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 67.73 |
| Shannon entropy | 0.59166 |
| G+C content | 0.62511 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -15.94 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975417 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 11591330 92 - 24543557 UUGUUGUAU--CCGCAACCGGAGUCGCCGAAAUGCCAUAUUCGUGGCCGUGGCCGUGGCCAUGACCCGUCGUCUGUGCUGCGCCGUGCAUUCUG ....(((((--.((((..((.((.((.((....(((((....)))))((((((....))))))...)).)).)).)).))))..)))))..... ( -30.70, z-score = -0.34, R) >droPer1.super_20 1839413 75 + 1872136 -----------AGGCGGCAUCCAUCGCCGAAAUGCCAUAUUCGUGGCCGUGGUCGUGGUCGUGG-CCAUGGCCAUGGCCAUGACCCG------- -----------.(((((......))))).....(((((....)))))((.((((((((((((((-(....)))))))))))))))))------- ( -44.10, z-score = -4.54, R) >dp4.chrXR_group6 3213801 81 + 13314419 -----------AGGCGGCAUCCAUCGCCGAAAUGCCAUAUUCGUGGCCGUGGUCGUGGUCGUGG-CCGUGGCCAUGGCCAUGGCCAUGACCCG- -----------.(((((......))))).....(((((....)))))((.((((((((((((((-((((....))))))))))))))))))))- ( -48.50, z-score = -4.40, R) >droAna3.scaffold_13337 5867004 82 - 23293914 CUGCCGGAG--UCAUGGCCGGAGUCGCCGAAAUGCCAUAUU------CGUGGCCGUGGCCAUGACCCGUCGUC----CUGUACUCCGUUCUCUG ..(.(((.(--(((((((((..(((((.(((........))------))))))..))))))))))))).)...----................. ( -31.10, z-score = -2.25, R) >droEre2.scaffold_4784 11578157 92 - 25762168 UUGUUGUAU--CCGCAACCGGAGUCGCCGAAAUGCCAUAUUCGUGGCCGUGGCCGUGGCCAUGACCCGUCGUCUGUGCUGCGCCGUGCAUUCUG ....(((((--.((((..((.((.((.((....(((((....)))))((((((....))))))...)).)).)).)).))))..)))))..... ( -30.70, z-score = -0.34, R) >droSim1.chr3L 10967275 94 - 22553184 UUGUUGUAUCCCCGCAACCGGAGUCGCCGAAAUGCCAUAUUCGUGGCCGUGGCCGUGGCCAUGACCCGUCGUCUGUGCUGCGCCGUGCAUUCUG ....(((((...((((..((.((.((.((....(((((....)))))((((((....))))))...)).)).)).)).))))..)))))..... ( -31.00, z-score = -0.54, R) >consensus UUGUUGUAU__CCGCAACCGGAGUCGCCGAAAUGCCAUAUUCGUGGCCGUGGCCGUGGCCAUGACCCGUCGUCUGUGCUGCGCCCUGCAUUCUG .............(((.........((((....(((((....)))))..))))((((((...(((.....)))...))))))...)))...... (-15.94 = -15.97 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:06 2011