
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,589,820 – 11,590,018 |
| Length | 198 |
| Max. P | 0.771849 |

| Location | 11,589,820 – 11,589,931 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.23466 |
| G+C content | 0.49588 |
| Mean single sequence MFE | -12.07 |
| Consensus MFE | -9.70 |
| Energy contribution | -10.96 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
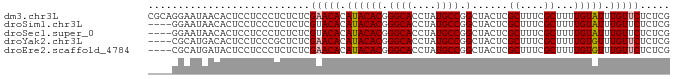
>dm3.chr3L 11589820 111 - 24543557 CCGGCUUAGUGCCGAAGGCUCCCCUCAACACCG-CCCCCCUUUUUUAUAACCUUACAUCUAGUGCUGAAAUCCCCUUGUAUCCUCCCUUUCGUCACUACUCAUCUCACUUUG .((((.....))))..(((.............)-)).......................(((((.(((((.................))))).))))).............. ( -14.75, z-score = -1.64, R) >droYak2.chr3L 11621017 91 - 24197627 CCGGCUUAGUGCCGAAUGCUCC------------ACCCCCCUUUUAAUAACCUUACAUCUAGUG---------CCUUGUUACCUCCCUUUCGUCACUACUCAUCUCAAUUCG .((((.....))))........------------.........................(((((---------.(................).))))).............. ( -8.69, z-score = -1.10, R) >droSec1.super_0 3779195 111 - 21120651 CCGGCUUAGUGCCGAAUGCUCCCCUCAUCUCUG-CCCCCCGUUUUUAUAACCUUACAUCUAGUGCUGAAAUCCCCUUGUUUCCUCCCUUUCGUCACUACUCAUCUCACUUUG .((((.....))))...................-.........................(((((.(((((.................))))).))))).............. ( -12.43, z-score = -1.71, R) >droSim1.chr3L 10965681 112 - 22553184 CCGGCUUAGUGCCGAAUGCUCCCCUCAUCUCUGCCCCCCCGUUUUUAUAACCUUACAUCUAGUGCUGAAAUCCCCUUGUUUCCUCCCUUUCGUCACUACUCAUCUCACUUUG .((((.....)))).............................................(((((.(((((.................))))).))))).............. ( -12.43, z-score = -1.66, R) >consensus CCGGCUUAGUGCCGAAUGCUCCCCUCAUCUCUG_CCCCCCGUUUUUAUAACCUUACAUCUAGUGCUGAAAUCCCCUUGUUUCCUCCCUUUCGUCACUACUCAUCUCACUUUG .((((.....)))).............................................(((((.(((((.................))))).))))).............. ( -9.70 = -10.96 + 1.25)


| Location | 11,589,931 – 11,590,018 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Shannon entropy | 0.10559 |
| G+C content | 0.52273 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11589931 87 + 24543557 CGAGAGAACAAAUACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUUCGAGAGAGGGAGGAGUGUUAUUCCUGCG .....(((((.((((....((....)).....((.((((....)))).)))))).))))).......(.((((((...)))))).). ( -27.10, z-score = -2.97, R) >droSim1.chr3L 10965793 83 + 22553184 CGAGAGAACAAAUACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUACGAGAGAGGGAGGAGUGUUAUUCC---- ...................((....))((((((((((((....))))((((......))))............).))))))).---- ( -18.50, z-score = -1.39, R) >droSec1.super_0 3779306 83 + 21120651 CGAGAGAACAAAUACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUACGAGAGAGGGAGGAGUGUUAUUCC---- ...................((....))((((((((((((....))))((((......))))............).))))))).---- ( -18.50, z-score = -1.39, R) >droYak2.chr3L 11621108 83 + 24197627 CGAGAGAACAAACACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUUCGAGAGCGGGAGGAGUGUCAUGCG---- ((((...(((.((((....((....))........((((....)))).)))).))).))))...(((.((......)).))).---- ( -23.70, z-score = -1.43, R) >droEre2.scaffold_4784 11576677 83 + 25762168 CGAGAGAACAAACACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUUCGAGAGAGGGAGGAGUAUCAUGCG---- ((((...(((.((((....((....))........((((....)))).)))).))).))))...(...((......))...).---- ( -21.10, z-score = -1.57, R) >consensus CGAGAGAACAAAUACAAAAGCGAAAGCGAGUAGCCGGCAUAGGUGCCCGUGUAUGUGUUCGAGAGAGGGAGGAGUGUUAUUCC____ ((((...(((.((((....((....))........((((....)))).)))).))).)))).......................... (-18.40 = -18.56 + 0.16)



| Location | 11,589,931 – 11,590,018 |
|---|---|
| Length | 87 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Shannon entropy | 0.10559 |
| G+C content | 0.52273 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
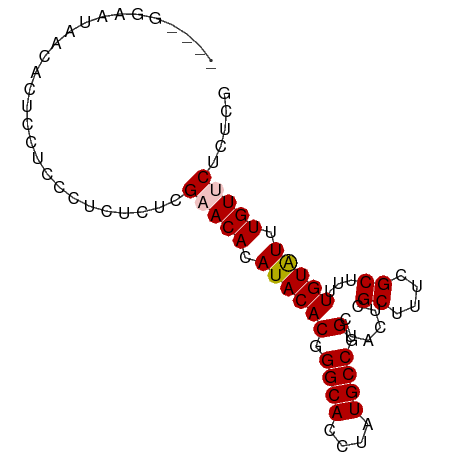
>dm3.chr3L 11589931 87 - 24543557 CGCAGGAAUAACACUCCUCCCUCUCUCGAACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUAUUUGUUCUCUCG .(.((((.......)))).).......(((((.((((((.((((....)))).)......((....))...))))).)))))..... ( -18.40, z-score = -2.25, R) >droSim1.chr3L 10965793 83 - 22553184 ----GGAAUAACACUCCUCCCUCUCUCGUACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUAUUUGUUCUCUCG ----(((.......)))..........(.(((.((((((.((((....)))).)......((....))...))))).))).)..... ( -13.10, z-score = -1.12, R) >droSec1.super_0 3779306 83 - 21120651 ----GGAAUAACACUCCUCCCUCUCUCGUACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUAUUUGUUCUCUCG ----(((.......)))..........(.(((.((((((.((((....)))).)......((....))...))))).))).)..... ( -13.10, z-score = -1.12, R) >droYak2.chr3L 11621108 83 - 24197627 ----CGCAUGACACUCCUCCCGCUCUCGAACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUGUUUGUUCUCUCG ----.((..((......))..))...((((((((....((((((....))))(((.....)))..))....))))))))........ ( -16.40, z-score = -0.93, R) >droEre2.scaffold_4784 11576677 83 - 25762168 ----CGCAUGAUACUCCUCCCUCUCUCGAACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUGUUUGUUCUCUCG ----......................((((((((....((((((....))))(((.....)))..))....))))))))........ ( -16.00, z-score = -1.55, R) >consensus ____GGAAUAACACUCCUCCCUCUCUCGAACACAUACACGGGCACCUAUGCCGGCUACUCGCUUUCGCUUUUGUAUUUGUUCUCUCG ...........................(((((.((((((.((((....)))).)......((....))...))))).)))))..... (-14.52 = -14.68 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:04 2011