| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,566,256 – 11,566,323 |
| Length | 67 |
| Max. P | 0.716531 |

| Location | 11,566,256 – 11,566,323 |
|---|---|
| Length | 67 |
| Sequences | 4 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.50303 |
| G+C content | 0.44889 |
| Mean single sequence MFE | -15.35 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
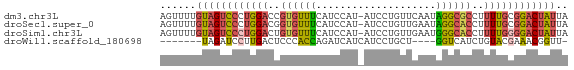
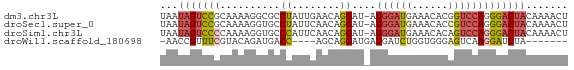
>dm3.chr3L 11566256 67 + 24543557 AGUUUUGUAGUCCCUGGACCGUGUUUCAUCCAU-AUCCUGUUCAAUAGGCGCCUUUUGCGGACUAUUA ......(((((((.((((..........)))).-..(((((...))))).((.....))))))))).. ( -12.90, z-score = -0.56, R) >droSec1.super_0 3758438 67 + 21120651 AGUUUUGUAGUCCCUGGACGGUGUUUCAUCCAU-AUCCUGUUGAAUAGGCACCUUUUGCGGACUAUUA ......(((((((.(..(.(((((((.((.((.-.......)).))))))))).)..).))))))).. ( -18.20, z-score = -1.88, R) >droSim1.chr3L 10941630 67 + 22553184 AGUUUUGUAGUCCCUGGACUGUGUUUCAUCCAU-AUCCUGUUGAAUGGGCACCUUUUGGGGACUAUUA ......((((((((..((..(((((.(((.((.-.......)).))))))))..))..)))))))).. ( -23.50, z-score = -2.96, R) >droWil1.scaffold_180698 2073893 56 + 11422946 -------UAGAUCCUUGACUCCCACCAGAUCAUCAUCCUGCU----GGUCAUCUGUACGAAACGGUU- -------..................(((((.((((......)----))).)))))............- ( -6.80, z-score = 1.26, R) >consensus AGUUUUGUAGUCCCUGGACUGUGUUUCAUCCAU_AUCCUGUUGAAUAGGCACCUUUUGCGGACUAUUA ......((((((((((((..((((((....................))))))..)))))))))))).. ( -9.16 = -9.85 + 0.69)
| Location | 11,566,256 – 11,566,323 |
|---|---|
| Length | 67 |
| Sequences | 4 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.50303 |
| G+C content | 0.44889 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -7.30 |
| Energy contribution | -8.67 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
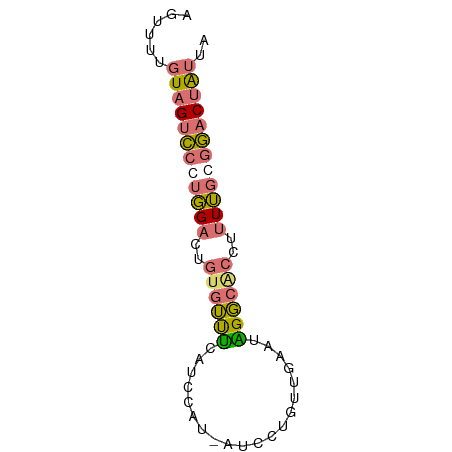
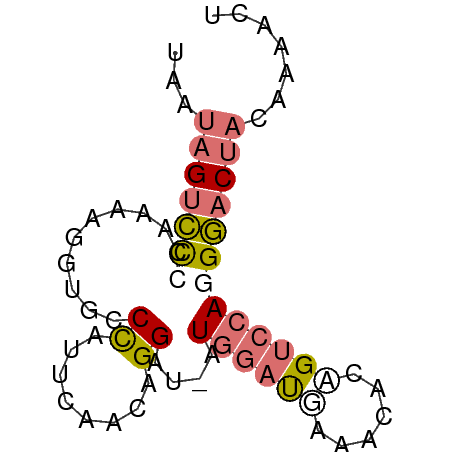
>dm3.chr3L 11566256 67 - 24543557 UAAUAGUCCGCAAAAGGCGCCUAUUGAACAGGAU-AUGGAUGAAACACGGUCCAGGGACUACAAAACU ...((((((.(((.((....)).)))....((((-.((.......))..))))..))))))....... ( -15.20, z-score = -1.65, R) >droSec1.super_0 3758438 67 - 21120651 UAAUAGUCCGCAAAAGGUGCCUAUUCAACAGGAU-AUGGAUGAAACACCGUCCAGGGACUACAAAACU ...(((((((((.....))).(((((....))))-)((((((......)))))).))))))....... ( -18.20, z-score = -2.71, R) >droSim1.chr3L 10941630 67 - 22553184 UAAUAGUCCCCAAAAGGUGCCCAUUCAACAGGAU-AUGGAUGAAACACAGUCCAGGGACUACAAAACU ...(((((((......(((..((((((.......-.))))))...)))......)))))))....... ( -18.10, z-score = -2.33, R) >droWil1.scaffold_180698 2073893 56 - 11422946 -AACCGUUUCGUACAGAUGACC----AGCAGGAUGAUGAUCUGGUGGGAGUCAAGGAUCUA------- -..((((.....)).(((..((----(.((((.......)))).)))..)))..)).....------- ( -14.20, z-score = -0.35, R) >consensus UAAUAGUCCCCAAAAGGUGCCCAUUCAACAGGAU_AUGGAUGAAACACAGUCCAGGGACUACAAAACU ...(((((((..........((........))....((((((......)))))))))))))....... ( -7.30 = -8.67 + 1.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:01 2011