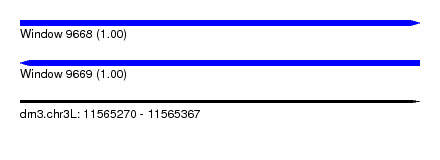
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,565,270 – 11,565,367 |
| Length | 97 |
| Max. P | 0.999096 |

| Location | 11,565,270 – 11,565,367 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 56.90 |
| Shannon entropy | 0.61741 |
| G+C content | 0.51444 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11565270 97 + 24543557 AUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUU-AGCCUAAGUCCAAAUAUAUUAAAAACUGUAAAAUCAGAGAGACUC ........((.(((.(((((((((.....))))))))).)))...))(((((-......)))))..............(((......)))........ ( -29.10, z-score = -1.99, R) >droSim1.chr3h_random 1352275 86 + 1452968 AAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUU-AGGCUAAGAAACCAUUGUAUUUAGAUCGCCAAAU----------- ...((((((..((.(((((((.(((...))).))))))).))....)).)))-)(((.......................)))....----------- ( -27.60, z-score = -0.88, R) >droPer1.super_1295 3577 95 + 6930 AUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUGGGCUUACAAAAGCCCAAACCACAAUUGUAAAAUAUAGAAGA--- ....(((((..(((.(((((((((.....))))))))).)))...(.((.(((((((((....))))))))))).)............)))))..--- ( -40.10, z-score = -3.91, R) >consensus AUAAAAACGCCGCUCCGCUGCAGGCGUAACCGGCAGCGCAGCUCCGCGGCUU_AGGCUAAGAAAAAAUAAAAUAAAAAUUGUAAAAU_____AGA___ .......(((.(((.(((((((((.....))))))))).)))...))).................................................. (-20.58 = -21.03 + 0.45)

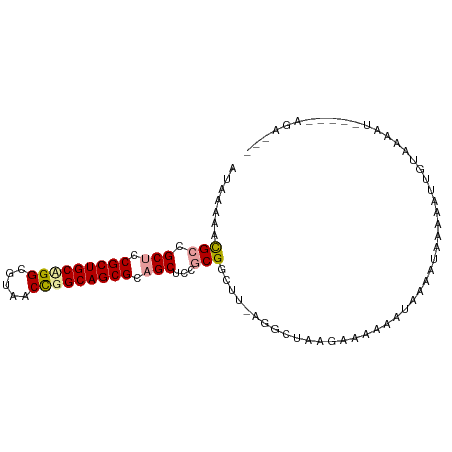
| Location | 11,565,270 – 11,565,367 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 56.90 |
| Shannon entropy | 0.61741 |
| G+C content | 0.51444 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -18.63 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11565270 97 - 24543557 GAGUCUCUCUGAUUUUACAGUUUUUAAUAUAUUUGGACUUAGGCU-AAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAU .(((((.((..(....................)..))...)))))-.....((((.(((.((((((.((.....)).)))))).))).))))...... ( -29.25, z-score = -0.39, R) >droSim1.chr3h_random 1352275 86 - 1452968 -----------AUUUGGCGAUCUAAAUACAAUGGUUUCUUAGCCU-AAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUU -----------....((((.((...........((((((..((..-..))..)))))).(((((((.(((...))).)))))))...))))))..... ( -32.80, z-score = -2.11, R) >droPer1.super_1295 3577 95 - 6930 ---UCUUCUAUAUUUUACAAUUGUGGUUUGGGCUUUUGUAAGCCCAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAU ---(((.((...........(((((((((((((((....)))))).))))))))).(((.(((((((.(.....).))))))).))))).)))..... ( -43.20, z-score = -3.59, R) >consensus ___UCU_____AUUUUACAAUUUUAAUUAAAUCUUUUCUUAGCCU_AAGCCGCGGAGCUGCGCUGCCGGCAACGCCAGCAGCGGAGCGGCGCUAAUAU .....................................................(..(((.((((((.((.....)).)))))).)))..)........ (-18.63 = -19.30 + 0.67)

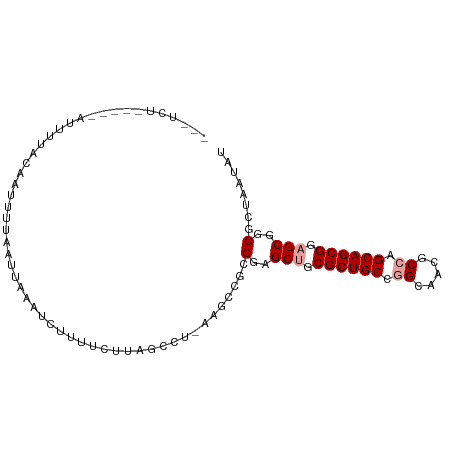
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:19:00 2011