| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,546,679 – 11,546,769 |
| Length | 90 |
| Max. P | 0.764674 |

| Location | 11,546,679 – 11,546,769 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 62.79 |
| Shannon entropy | 0.67529 |
| G+C content | 0.50327 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -10.05 |
| Energy contribution | -9.03 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11546679 90 - 24543557 ---AGUAGUAGAUUCUGCUACUAUUC-UACUGCCACUUUGCCGUGUUCUCUUGUUACCGAGCUGUGUGGCAACGCCAGC-GCAGUCAGCUGCUGA------- ---((((((((...))))))))....-...((((((...((((.((.........)))).))...))))))....((((-((.....)).)))).------- ( -29.00, z-score = -1.13, R) >droSim1.chr3L 10930596 92 - 22553184 AAAAGUAGUAGAUUCUGCUACUAUUC-UACGGCCACUUGGCCAAGUUCUCUUGUUACCGA-CCAUGUGGCAACGCCAGC-GCAGUCAGCUGCUGA------- ...((((((((...))))))))....-....(((((.((((((((....)))).....).-))).))))).....((((-((.....)).)))).------- ( -28.60, z-score = -1.47, R) >droSec1.super_0 3746441 89 - 21120651 ---AGUAGUAGAUUCUGCUACUAUUC-UACUGCCACUUGGCCGAGUUCUCUUGUUACCGA-CCAUGUGGCAACGCCAGC-GCAGUCAGCUGCUGA------- ---((((((((...))))))))....-...((((((.(((.((.((.........)))).-))).))))))....((((-((.....)).)))).------- ( -30.60, z-score = -2.08, R) >droYak2.chr3L 11581075 76 - 24197627 --AAGUAGUAGAUUCUGCUACUCUUU-UACUGCCA--------------CUUGUUACCGA-CCAUGUGGCAACGCCAGC-GCAGUCAGCUGCUGA------- --.((((((((...))))))))....-...(((((--------------(.((.......-.)).))))))....((((-((.....)).)))).------- ( -24.30, z-score = -1.89, R) >droEre2.scaffold_4784 11546824 74 - 25762168 ----GUAGUAGAUUCUGCUACAGUUC-UACUGCCA--------------CUUGUUACCGA-CCAUGUGGCAACGCCAGC-GCAGUCAGCUGCUGA------- ----(((((((...))))))).....-...(((((--------------(.((.......-.)).))))))....((((-((.....)).)))).------- ( -24.00, z-score = -1.23, R) >dp4.chrXR_group6 3188323 81 + 13314419 ---CGCAGUAGAUUCUACUGCUAUUUGCGCAGUGA--------GGUCCGUUUCUUGUCG--UCUGGCAACGCUGCACAG-GCCGUCAGCUGCUGA------- ---.(((((.(((..((((((.......)))))).--------.))).......((.((--(((((((....))).)))-).)).)))))))...------- ( -28.80, z-score = -0.83, R) >droPer1.super_20 1813575 81 + 1872136 ---CGCAGUAGAUUCUACUGCUAUUUGCGCAGUGA--------AGUCCGUUUCUUGUCG--UCUGGCAACGCUGCACAG-GCCGUCAGCUGCUGA------- ---.(((((.((((.((((((.......)))))).--------)))).......((.((--(((((((....))).)))-).)).)))))))...------- ( -27.50, z-score = -0.72, R) >droVir3.scaffold_13049 23765693 80 - 25233164 ---AGUACUAAAUUCUACUACUUAUUACAUUGCCC-------GUGAAGCGGUUUAGUAG--UCUGGCAACGCUGU---CUGCUGUCAGCUGCUAA------- ---((((........))))..............((-------(.....))).(((((((--.((((((..((...---..)))))))))))))))------- ( -19.60, z-score = -0.38, R) >droGri2.scaffold_15110 10662685 89 + 24565398 ---AGUAGUAAAUUCUACUACUAUUUUCA-UGUGA-------ACUAAAUGUUCAUGCCA--UCUGGCAACGCUACCCGCUGCUGUCAGCUGUCAGCUGCUGA ---(((((((.....))))))).......-.((((-------((.....))))))(((.--...)))..........((.((((.(....).)))).))... ( -25.70, z-score = -2.60, R) >consensus ___AGUAGUAGAUUCUGCUACUAUUU_UACUGCCA________AGUUCUCUUGUUACCGA_CCAUGUGGCAACGCCAGC_GCAGUCAGCUGCUGA_______ ....(((((((...)))))))............................................(((....))).....((.(....).)).......... (-10.05 = -9.03 + -1.02)

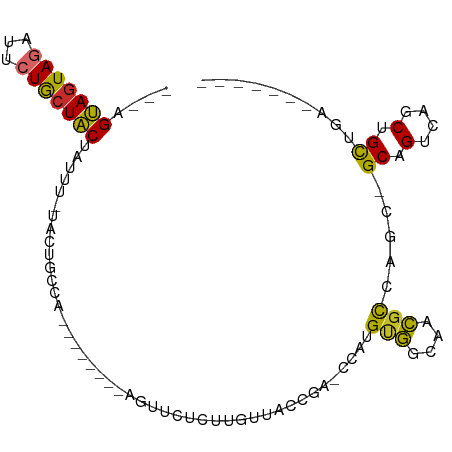

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:57 2011