
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,534,910 – 11,535,012 |
| Length | 102 |
| Max. P | 0.538391 |

| Location | 11,534,910 – 11,535,012 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Shannon entropy | 0.38662 |
| G+C content | 0.56975 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
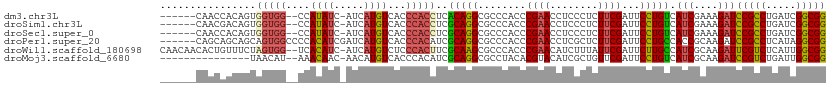
>dm3.chr3L 11534910 102 + 24543557 CCGCCGAUCAGGCGGAUCUUUCGAUGACAGGAAUCGAAGAGGGAGGUUCGGGUGGGCGCCUGUGAGGUGGGUGACAUGAU-GAUAUGG--CCACCACUGUGGUUG------ (((((.....)))))......((((.(((((((((.........))))).(((((.((((.....))))(....).....-.......--))))).)))).))))------ ( -37.80, z-score = -1.27, R) >droSim1.chr3L 10917798 102 + 22553184 CCGCCGAUCAGGCGGAUCUUUCGAUGACAGGAAUCGAAGAGGGAGGUUCGGGUGGGCGCCUGCGAGGUGGGUGACAUGAU-GAUAUGG--CCACCACUGUCGUUG------ (((((.....)))))......((((((((((((((.........))))).(((((.((((..(...)..)))).((((..-..)))).--))))).)))))))))------ ( -42.30, z-score = -2.21, R) >droSec1.super_0 3734266 102 + 21120651 CCGCCGAUCAGGCGGAUCUUUCGAUGACAGGAAUCGAAGAGGGAGGUUCGGGUGGGCGCCUGCGAGGUGGGUGACAUGAU-GAUAUGG--CCACCACUGUGGUUG------ (((((.....)))))......((((.(((((((((.........))))).(((((.((((..(...)..)))).((((..-..)))).--))))).)))).))))------ ( -38.90, z-score = -1.42, R) >droPer1.super_20 1803365 105 - 1872136 CCGCCUAUGAGGCGGAUCUUGCGGUGGCAGGAAUCGAAGAGCGAGGUUCGGGUGGGCGCCUGCGAUGUGGGUGACAUGAUCGAUGUGGGGCCACUGCUGCUGCUG------ (((((.....))))).....((((..(((((((((.........))))).....(((.((..((((...(....)...))))....)).))).))))..))))..------ ( -46.90, z-score = -2.34, R) >droWil1.scaffold_180698 2045540 108 + 11422946 CCGCCAAUGAGACGAAUCUUGCGAUGGCAAGAAUCGAAUAAAGAUGUUCGGGUGGGCGCUUGCGAAGUGGGAGACAUGAU-GAUGUGA--CCACUAGAAACAGUGUUGUUG (((((........((.((((((....)))))).))(((((....))))).)))))(((((..(..(((((...((((...-.))))..--))))).)....)))))..... ( -33.30, z-score = -2.37, R) >droMoj3.scaffold_6680 18376062 93 + 24764193 CCGCCAAUCAGACGGAUCUUGCGAUGACAGGAAUCGAACAGCGAUGUACGUGUAGGCGCCUGCGAUGUGGGUGACAUGUU-GUUGUUU--AUGUUA--------------- (((.(.....).))).(((((......)))))...((((((((((((((((((((....)))).)))).(....))))))-)))))))--......--------------- ( -25.10, z-score = 0.59, R) >consensus CCGCCGAUCAGGCGGAUCUUGCGAUGACAGGAAUCGAAGAGGGAGGUUCGGGUGGGCGCCUGCGAGGUGGGUGACAUGAU_GAUAUGG__CCACCACUGUCGUUG______ (((((.....))))).((((.((((.......))))))))..........(((((.((((..(...)..)))).((((.....))))...)))))................ (-22.17 = -22.28 + 0.12)

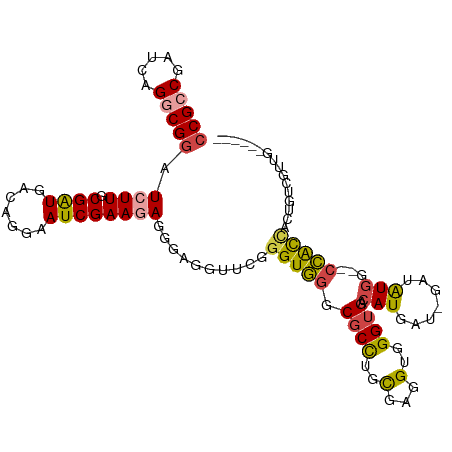

| Location | 11,534,910 – 11,535,012 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Shannon entropy | 0.38662 |
| G+C content | 0.56975 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11534910 102 - 24543557 ------CAACCACAGUGGUGG--CCAUAUC-AUCAUGUCACCCACCUCACAGGCGCCCACCCGAACCUCCCUCUUCGAUUCCUGUCAUCGAAAGAUCCGCCUGAUCGGCGG ------...((...(.(((((--..((((.-...))))...))))).).((((((......((((........)))).........(((....))).))))))...))... ( -25.40, z-score = -0.78, R) >droSim1.chr3L 10917798 102 - 22553184 ------CAACGACAGUGGUGG--CCAUAUC-AUCAUGUCACCCACCUCGCAGGCGCCCACCCGAACCUCCCUCUUCGAUUCCUGUCAUCGAAAGAUCCGCCUGAUCGGCGG ------....((((.((((((--.....))-))))))))..((.((...((((((......((((........)))).........(((....))).))))))...)).)) ( -29.60, z-score = -1.42, R) >droSec1.super_0 3734266 102 - 21120651 ------CAACCACAGUGGUGG--CCAUAUC-AUCAUGUCACCCACCUCGCAGGCGCCCACCCGAACCUCCCUCUUCGAUUCCUGUCAUCGAAAGAUCCGCCUGAUCGGCGG ------...((...(.(((((--(.((...-...))))))))).((...((((((......((((........)))).........(((....))).))))))...)).)) ( -25.90, z-score = -0.62, R) >droPer1.super_20 1803365 105 + 1872136 ------CAGCAGCAGCAGUGGCCCCACAUCGAUCAUGUCACCCACAUCGCAGGCGCCCACCCGAACCUCGCUCUUCGAUUCCUGCCACCGCAAGAUCCGCCUCAUAGGCGG ------........((.(((((.....(((((..((((.....)))).(((((..(......)..))).))...)))))....))))).)).....(((((.....))))) ( -28.90, z-score = -1.19, R) >droWil1.scaffold_180698 2045540 108 - 11422946 CAACAACACUGUUUCUAGUGG--UCACAUC-AUCAUGUCUCCCACUUCGCAAGCGCCCACCCGAACAUCUUUAUUCGAUUCUUGCCAUCGCAAGAUUCGUCUCAUUGGCGG .........(((....(((((--..((((.-...))))...)))))..)))..((((.....(((........)))((.((((((....)))))).))........)))). ( -23.90, z-score = -2.30, R) >droMoj3.scaffold_6680 18376062 93 - 24764193 ---------------UAACAU--AAACAAC-AACAUGUCACCCACAUCGCAGGCGCCUACACGUACAUCGCUGUUCGAUUCCUGUCAUCGCAAGAUCCGUCUGAUUGGCGG ---------------......--.......-.................(((((..(..(((((.....)).)))..)...))))).(((....)))(((((.....))))) ( -17.50, z-score = 0.32, R) >consensus ______CAACCACAGUAGUGG__CCACAUC_AUCAUGUCACCCACCUCGCAGGCGCCCACCCGAACCUCCCUCUUCGAUUCCUGUCAUCGAAAGAUCCGCCUGAUCGGCGG ................(((((....((((.....))))...)))))..(((((........((((........))))...))))).(((....)))(((((.....))))) (-18.55 = -18.63 + 0.09)

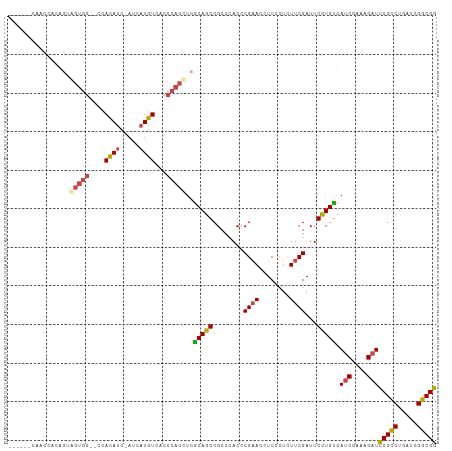
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:55 2011