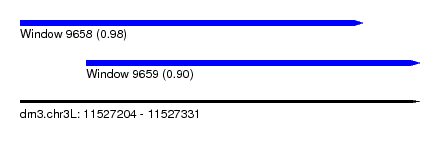
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,527,204 – 11,527,331 |
| Length | 127 |
| Max. P | 0.982184 |

| Location | 11,527,204 – 11,527,313 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 60.88 |
| Shannon entropy | 0.74699 |
| G+C content | 0.34770 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -6.42 |
| Energy contribution | -8.47 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
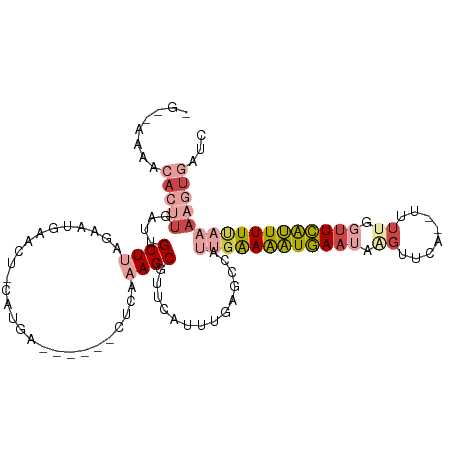
>dm3.chr3L 11527204 109 + 24543557 CUUUAAAAAACUUAAUUGCUUAGAAUGAACUGCAUGAGUUAGCCUUCAAGCGUUCAUUUGAGCCAUUGAAAAUGAAUAAGUUCA--UUUUGGUUCAUUUUUAAAACUGAUC .((((((((.............((((((((.((.((((......)))).))))))))))((((((....(((((((....))))--))))))))).))))))))....... ( -26.70, z-score = -2.91, R) >droEre2.scaffold_4784 11527602 86 + 25762168 GGGCAAAACACAUAACUGCUU-----------------------UCGAAGCGUUCAUUUUAGCCUUCGGAAAUGAAUAGGUUCA--UUUUGGUUCAUUUUUAAAAGUGAUC ........(((..........-----------------------.(((((.(((......))))))))(((((((((..(....--..)..))))))))).....)))... ( -15.60, z-score = -0.48, R) >droYak2.chr3L 11561212 98 + 24197627 GGCCAAAACACUUGAUUGCUUAGAAUGAACG-----------UGUCACAGCGUUCAUUUUAGCCUCUGAAAAUGAACAAGUUCA--UUUGGCUUCAUUUUCAAAAGUGAUC (((((((..(((((...(((.((((((((((-----------(......)))))))))))))).((.......)).)))))...--)))))))((((((....)))))).. ( -29.80, z-score = -3.61, R) >droSec1.super_0 3726801 102 + 21120651 -GUUAUAACACUUGAUUGCUUAGAAUGAACU-CAUGAA-----CUCAAAGCGUUCCUUUGAGCCAUUGAAAAUGAAUGAUUUCA--UUUUGGUUCAUUUUUAAAAGUGAUC -............(((..(((((((((((((-((....-----(((((((.....)))))))....))((((((((....))))--))))))))))))))...)))..))) ( -26.90, z-score = -3.40, R) >droSim1.chr3L 10909919 99 + 22553184 ----AUAACACUUGAAUGCUUAGAAUGAACU-CAUGAA-----UUCAAAGCGUUCCUUUGAGCCAUUGAAAAUGAAUGAGUUCA--UUUUGGUUCAUUUUUAAAAGUGAUC ----....((((((((((.((((((((((((-(((...-----(((((.(.((((....))))).))))).....)))))))))--))))))..)))))....)))))... ( -29.20, z-score = -4.13, R) >droMoj3.scaffold_6680 18367160 86 + 24764193 -----------------GCUUUGAUUCAGAUUCAGGUUGACUGAAUGCAGCGCUCAGCUGCAAAUUCAAAGUUCACAGAGCAAAGCUUUCAAUUGGGUUUCAU-------- -----------------((((((......((((((.....))))))(((((.....)))))..............)))))).((((((......))))))...-------- ( -25.40, z-score = -1.57, R) >consensus _G__AAAACACUUGAUUGCUUAGAAUGAACU_CAUGA______CUCAAAGCGUUCAUUUGAGCCAUUGAAAAUGAAUAAGUUCA__UUUUGGUUCAUUUUUAAAAGUGAUC ........(((((....((((.((....................)).))))..............((((((((((((.((........)).)))))))))))))))))... ( -6.42 = -8.47 + 2.05)

| Location | 11,527,225 – 11,527,331 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Shannon entropy | 0.39483 |
| G+C content | 0.32597 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -11.28 |
| Energy contribution | -13.72 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
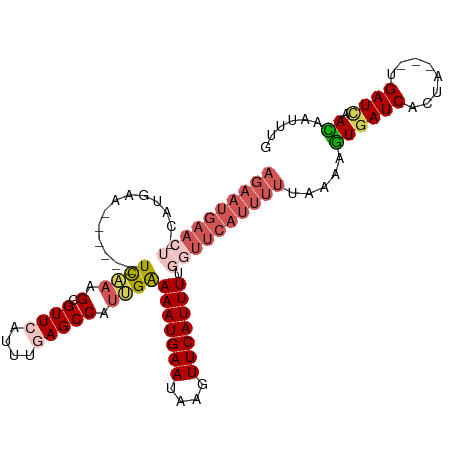
>dm3.chr3L 11527225 106 + 24543557 AGAAUGAACUGCAUGAGUUAGCCUUCAAGCGUUCAUUUGAGCCAUUGAAAAUGAAUAAGUUCAUUUUGGUUCAUUUUUAAAACUGAUCACUA---UGAUCAAGAAUUUG .((((((((.((.((((......)))).))))))))))((((((....(((((((....)))))))))))))...........(((((....---.)))))........ ( -28.20, z-score = -2.92, R) >droEre2.scaffold_4784 11527623 86 + 25762168 -----------------------UCGAAGCGUUCAUUUUAGCCUUCGGAAAUGAAUAGGUUCAUUUUGGUUCAUUUUUAAAAGUGAUCACCAGACUGAUGAACAAUUUG -----------------------((((((.(((......)))))))))..........((((((((((((((((((....))))))..)))))...)))))))...... ( -20.80, z-score = -2.00, R) >droYak2.chr3L 11561233 98 + 24197627 AGAAUGAACG-----UG------UCACAGCGUUCAUUUUAGCCUCUGAAAAUGAACAAGUUCAUUUGGCUUCAUUUUCAAAAGUGAUCACAAGACUGAUCCACAAUUUG ((((((((((-----(.------.....)))))))))))......(((((((((.((((....))))...)))))))))...(((((((......)))).)))...... ( -26.40, z-score = -2.74, R) >droSec1.super_0 3726821 99 + 21120651 AGAAUGAACU-CAUGAA-----CUCAAAGCGUUCCUUUGAGCCAUUGAAAAUGAAUGAUUUCAUUUUGGUUCAUUUUUAAAAGUGAUCACUA----GAUUAAUAAUUUG ((((((((((-((....-----(((((((.....)))))))....))((((((((....))))))))))))))))))......(((((....----)))))........ ( -22.50, z-score = -2.27, R) >droSim1.chr3L 10909936 100 + 22553184 AGAAUGAACU-CAUGAA-----UUCAAAGCGUUCCUUUGAGCCAUUGAAAAUGAAUGAGUUCAUUUUGGUUCAUUUUUAAAAGUGAUCACUA---UGAUCAAUAAUUUG ((((((((((-(((...-----(((((.(.((((....))))).))))).....)))))))))))))................(((((....---.)))))........ ( -25.90, z-score = -2.80, R) >consensus AGAAUGAACU_CAUGAA______UCAAAGCGUUCAUUUGAGCCAUUGAAAAUGAAUAAGUUCAUUUUGGUUCAUUUUUAAAAGUGAUCACUA___UGAUCAACAAUUUG ((((((((((.............((((.(.((((....))))).))))(((((((....))))))).)))))))))).....(((((((......))))).))...... (-11.28 = -13.72 + 2.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:52 2011