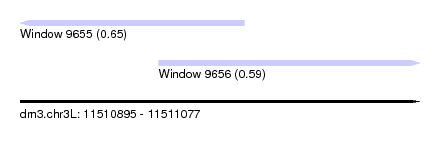
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,510,895 – 11,511,077 |
| Length | 182 |
| Max. P | 0.650977 |

| Location | 11,510,895 – 11,510,997 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Shannon entropy | 0.53766 |
| G+C content | 0.53286 |
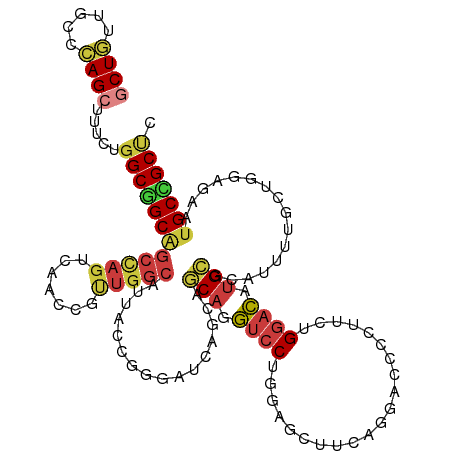
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
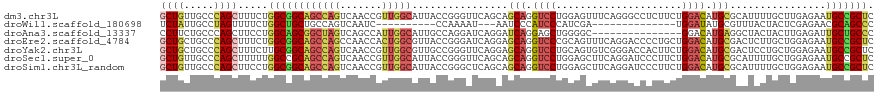
>dm3.chr3L 11510895 102 - 24543557 AACGGUUGACUGGCU-GCCGCCAGAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAAC---AGAUUCAUAUUCAGAGCUGGU-------- ..(((((..(((((.-...)))))..))))).....((((..((((.((((((........))))))(((.......)))))---))..((.......))))))..-------- ( -31.30, z-score = -0.61, R) >droMoj3.scaffold_6680 6877279 95 + 24764193 -CGGGUUGGCUGGCC-GCAGCCAGGAAACUGGGCAGCAGCACCUGCCGCUGGUACAGGUCGAGCAGCUGCUUGAACUGCAGC---AGCUUCGUCUCCGGC-------------- -(((....((..((.-((((((((....))))((....))..)))).))..))...((.(((((.(((((.......)))))---.)).))).)))))..-------------- ( -42.80, z-score = -0.26, R) >droGri2.scaffold_15110 14365084 93 - 24565398 ----------UGGCCAGCGGCCAGGAAGCUGGGCAGCAGCACUUGUCGCUGAUACAAAUCAAGUAGUUGCUUAAAUUGCAUU---AGUUUUAUACCCGUUUCCGUU-------- ----------(((((...)))))((((((.(((((((.((....)).))))...........((((((.....))))))...---.........)))))))))...-------- ( -30.40, z-score = -1.91, R) >droVir3.scaffold_13049 1957321 104 + 25233164 -CUGGCUGGCUGGCC-GCGGCCAGAAAACUGGGCAGCAGCACCUGGCGCUGGUAUAAAUCCAGCAGCUGCUUAAACUGCAUUUGCAGCUUUAGACCCGACUCCGGC-------- -...((((((((((.-...))))).....((((((((.((.....))(((((.......))))).)))))(((((((((....)))).)))))..)))...)))))-------- ( -43.10, z-score = -1.17, R) >droWil1.scaffold_180698 9415485 105 + 11422946 ---GAUUGACUGGCA-GCAGCCAGAAAACUAGGCAAUAGAACUUGCCGCUGAUAUAGAUCGAUAAGUUGUUUAAAUUGUACA---AGAUUAAGACCAUUUCCUUGAUUUGCU-- ---......(((((.-...))))).......(((((......))))).......((((((((.((((.((((.((((.....---.)))).)))).))))..))))))))..-- ( -23.50, z-score = -1.48, R) >droPer1.super_20 1773515 108 + 1872136 AUCGGCUGGCUGGCC-GCGGCCAGGAAGCUGGGCAGGAGCACCUGCCUCUGGUACAGGUCGAUCAGCUGUUUGAACUGCAGC---AGAUUCAGAUUCAGACCCGAUCCUGAC-- ..((((((..(((((-...(((((......(((((((....))))))))))))...)))))..))))))(((((((((...)---)).)))))).((((........)))).-- ( -44.90, z-score = -1.33, R) >dp4.chrXR_group6 10167969 110 - 13314419 AUCGGCUGGCUGGCC-GCGGCCAGGAAGCUGGGCAGGAGCACCUGCCUCUGGUACAGGUCGAUCAGCUGUUUGAACUGCAGC---AGAUUCAGAUUCAGACCCGAUUCCGAUUC ((((((((..(((((-...(((((......(((((((....))))))))))))...)))))..)))).((((((((((.(..---...).))).))))))).))))........ ( -44.50, z-score = -1.32, R) >droAna3.scaffold_13337 5796277 102 - 23293914 AAUGGCUGACUAGCC-GCUGCCAGGAAGCUGGGCAGAAGGACCUGUCUCUGAUACAGAUCGAUGAGUUGCUUGAAUUGCAAA---AGAUUCAUAUUCAGGGCAGGU-------- ...((((....))))-.(((((((....)).)))))....(((((((.((((((..((((......((((.......)))).---.))))..)).)))))))))))-------- ( -35.70, z-score = -1.84, R) >droEre2.scaffold_4784 11512394 102 - 25762168 AGUGGUUGGCUGGCU-GCCGCCAGAAAGCUGGGCAGCAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAGC---AGAUUCAUAUUCAAAGCUGGU-------- ..((((((((((.((-(((..(((....))))))))))))(((.......)))......))))))(((((.......)))))---.....................-------- ( -33.00, z-score = 0.62, R) >droYak2.chr3L 11545039 102 - 24197627 AACGGUUGACUGGCU-GCCGCAAGAAAGCUGGGCAGCAGCACCUGCCGCUGGUACAAAUCGAUCAGCUGCUUGAACUGCAGC---AGAUUCAUAUUCAACGCUGGU-------- ....((((((..(((-..(....)..)))..).((((.((....)).)))).........((((.(((((.......)))))---.)))).....)))))......-------- ( -33.80, z-score = -0.18, R) >droSec1.super_0 3709267 102 - 21120651 AACGGUUGACUGGCU-GCGGCCAAAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAAC---AGAUUCAUAUUCAGAGCUGGU-------- ..(((((..((((((-(..(((.........)))..))))..((((.((((((........))))))(((.......)))))---)).........))))))))..-------- ( -29.90, z-score = -0.46, R) >droSim1.chr3L_random 527011 102 - 1049610 AACGGUUGACUGGCU-GCCGCCAGGAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAAC---AGAUUCAUAUUCAGAGCUGGU-------- ..(((((..(((((.-...)))))..))))).....((((..((((.((((((........))))))(((.......)))))---))..((.......))))))..-------- ( -33.30, z-score = -0.93, R) >consensus AACGGUUGACUGGCC_GCGGCCAGAAAGCUGGGCAGCAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAGC___AGAUUCAUAUUCAGAGCUGGU________ ..(((((..(((((.....)))))..)))))(((((......)))))((((((........))))))(((.......))).................................. (-20.56 = -21.38 + 0.82)



| Location | 11,510,958 – 11,511,077 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.54928 |
| G+C content | 0.58686 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.67 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11510958 119 + 24543557 GCUGUUGCCCAGCUUUCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGUUCAGCAGCAGGUCCUGGAGUUUCAGGGCCUCUUCUGGACAUGCGCAUUUUGCUUGAGAAUGCCGCUC (((((((((.((....)))))))))))............((((((..(((((((((.((.(((((((((....))))))))))).))))))....((.....)))))..)))))).... ( -48.90, z-score = -1.64, R) >droWil1.scaffold_180698 9415554 92 - 11422946 UCUAUUGCCUAGUUUUCUGGCUGCUGCCAGUCAAUC----------CCAAAAU---AAUCCCAUCCCAUCGA--------------UGGAUAUGCGUUUACUACUCGAGAACGCAGCCC ..((((.....(((..(((((....)))))..))).----------....)))---)...(((((.....))--------------)))...(((((((.(.....).))))))).... ( -22.60, z-score = -2.57, R) >droAna3.scaffold_13337 5796340 104 + 23293914 CCUUCUGCCCAGCUUCCUGGCAGCGGCUAGUCAGCCAUUGGCAUUGCCAGGAUCAGGAUCAGGAGCUGGGGC---------------GGACAUGAGGCUACUACUUGAGAUUGCUGCCC ...((((((((((((((((((((..((((((.....)))))).)))))))(((....))).)))))).))))---------------))).....(((..(...........)..))). ( -40.60, z-score = -0.09, R) >droEre2.scaffold_4784 11512457 119 + 25762168 GCUGCUGCCCAGCUUUCUGGCGGCAGCCAGCCAACCACUGGCGUUACCGGGAUCAGGAGCAGGUCCCGCAGUUUCAGGACCCCUGCUGGACAUGCGACUCUUGCUGGAGAAUGCCGCUC (((((((((.((....)))))))))))............((((((.((((.(..((..(((.((((.((((...........)))).)))).)))..))..).))))..)))))).... ( -50.80, z-score = -1.07, R) >droYak2.chr3L 11545102 119 + 24197627 GCUGCUGCCCAGCUUUCUUGCGGCAGCCAGUCAACCGUUGGCGUUGCCGGGUUCAGGAGCAGGUCCUGCAGUGUCGGGACCACUUCUGGACAUGCGACUCCUGCUGGAGAAUGCCGCUC ((((.....))))......((((((.(((((........((.(((((...(((((((((..(((((((......))))))).)))))))))..))))).)).)))))....)))))).. ( -54.70, z-score = -1.80, R) >droSec1.super_0 3709330 119 + 21120651 GCUGUUGCCCAGCUUUUUGGCCGCAGCCAGUCAACCGUUGGCAUUACCGGGUUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUC ((.((..((((((......((.((((((((.(....))))))........((((((.((..((((((((....)))))))).)).)))))).))))).....))))).)...)).)).. ( -42.70, z-score = -0.16, R) >droSim1.chr3L_random 527074 119 + 1049610 GCUGUUGCCCAGCUUCCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGCUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUC ((((.....)))).....(((((((.((((.(((....((((((..(((((((....))).((((((((....))))))))....))))..)))).))..)))))))....))))))). ( -46.10, z-score = -0.19, R) >consensus GCUGUUGCCCAGCUUUCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGAUCAGCAGCAGGUCCUGGAGCUUCAGGACCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUC (((((((((.((....)))))))))))............((((((..(.((.......(((.((((.....................)))).)))........)).)..)))))).... (-22.20 = -23.67 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:49 2011