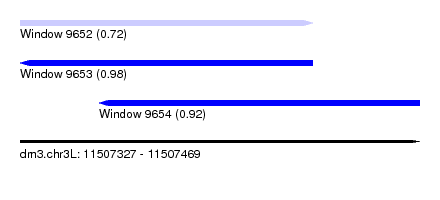
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,507,327 – 11,507,469 |
| Length | 142 |
| Max. P | 0.982961 |

| Location | 11,507,327 – 11,507,431 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Shannon entropy | 0.28831 |
| G+C content | 0.55522 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -27.73 |
| Energy contribution | -28.79 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11507327 104 + 24543557 GGAACUUCAGGUCCUCUUAAUUGGUGGGACCCCCCCCAUUCAGGCAAACCCUUUGACAACAACCACAACGAAGCAUUGUACUUG-UUUGCCUGGCACGGGGGAUC .........((((((((.....)).))))))(((((...((((((((((......((((................))))....)-)))))))))...)))))... ( -34.89, z-score = -1.35, R) >droSim1.chr3L 10892097 103 + 22553184 GGAGCCUCAGCUCCUCUUAAUUGGUGGGACCCCCC--AUUCAGGCAAACCCUUUGACAACAACCACAACGAAGCAUUGUACUUGGUUUGCCUGGCACGGGGGAUC (((((....)))))(((((.....)))))(((((.--..(((((((((((.....((((................))))....)))))))))))...)))))... ( -38.29, z-score = -2.33, R) >droSec1.super_0 3705640 103 + 21120651 GGAGCCUCAGCUCCUCUUAAUUGGUGGGACCCCCC--AUUCAGGCAAACCCUUUGACAACAACCACAACGAAGCAUUGUACUUGGUUUGCCUGGCACGGGGGAUC (((((....)))))(((((.....)))))(((((.--..(((((((((((.....((((................))))....)))))))))))...)))))... ( -38.29, z-score = -2.33, R) >droEre2.scaffold_4784 11508759 86 + 25762168 GGAGCUCCAGAUCCUCUUAAUUGGUGGGACCACCC--AUUACGCCGAGCCCUUUGACAAC-------------CAUUGUACUCG--GUGGCGGGCAGGGCGGG-- (((........))).......(((((((....)))--))))((((..((((........(-------------(((((....))--)))).))))..))))..-- ( -28.80, z-score = 0.14, R) >consensus GGAGCCUCAGCUCCUCUUAAUUGGUGGGACCCCCC__AUUCAGGCAAACCCUUUGACAACAACCACAACGAAGCAUUGUACUUG_UUUGCCUGGCACGGGGGAUC (((((....)))))(((((.....)))))(((((.....(((((((((((.....((((................))))....)))))))))))...)))))... (-27.73 = -28.79 + 1.06)

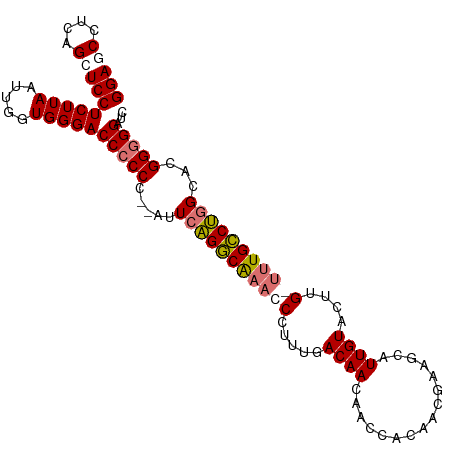
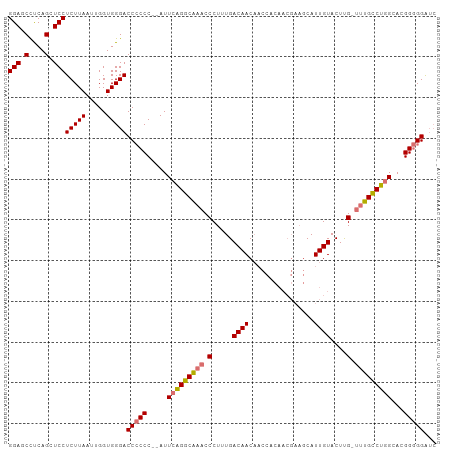
| Location | 11,507,327 – 11,507,431 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Shannon entropy | 0.28831 |
| G+C content | 0.55522 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -29.31 |
| Energy contribution | -31.75 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11507327 104 - 24543557 GAUCCCCCGUGCCAGGCAAA-CAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAUGGGGGGGGUCCCACCAAUUAAGAGGACCUGAAGUUCC ...(((((((..((((((((-(...(((((((...))))))).(((....)))...))))))))).)))))))(((((((.........).))))))........ ( -41.80, z-score = -2.42, R) >droSim1.chr3L 10892097 103 - 22553184 GAUCCCCCGUGCCAGGCAAACCAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAU--GGGGGGUCCCACCAAUUAAGAGGAGCUGAGGCUCC ((((((((((..((((((((((...(((((((...))))))).(((....)))..)))))))))).))--)))))))).............(((((....))))) ( -48.90, z-score = -4.53, R) >droSec1.super_0 3705640 103 - 21120651 GAUCCCCCGUGCCAGGCAAACCAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAU--GGGGGGUCCCACCAAUUAAGAGGAGCUGAGGCUCC ((((((((((..((((((((((...(((((((...))))))).(((....)))..)))))))))).))--)))))))).............(((((....))))) ( -48.90, z-score = -4.53, R) >droEre2.scaffold_4784 11508759 86 - 25762168 --CCCGCCCUGCCCGCCAC--CGAGUACAAUG-------------GUUGUCAAAGGGCUCGGCGUAAU--GGGUGGUCCCACCAAUUAAGAGGAUCUGGAGCUCC --..((((..((((((.((--((.......))-------------)).))....))))..))))((((--.((((....)))).))))...(((.(....).))) ( -26.00, z-score = 0.67, R) >consensus GAUCCCCCGUGCCAGGCAAA_CAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAU__GGGGGGUCCCACCAAUUAAGAGGAGCUGAAGCUCC ((((((((....((((((((((....((((((((.......)).)))))).....)))))))))).....)))))))).............(((((....))))) (-29.31 = -31.75 + 2.44)

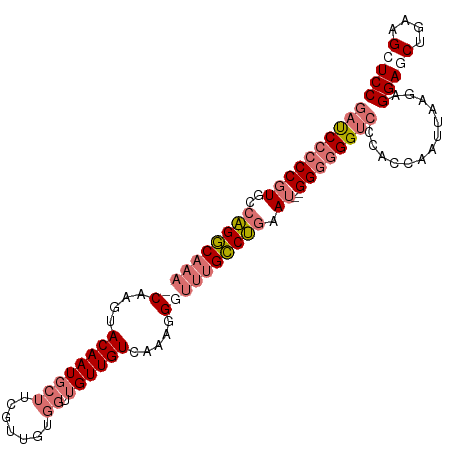
| Location | 11,507,355 – 11,507,469 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.42999 |
| G+C content | 0.53627 |
| Mean single sequence MFE | -43.29 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11507355 114 - 24543557 UGGCAACUCCGCUUACCCAAG-GGGCAUAUAAGUGGAUCGAUCCCCCGUGCCA-GGCAAA-CAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAUGGGGGGGGU .(....)(((((((((((...-)))....))))))))....((((((((..((-((((((-(...(((((((...))))))).(((....)))...))))))))).))))))))... ( -45.40, z-score = -1.75, R) >droSim1.chr3L 10892125 113 - 22553184 UGGCAACUCCGCUUACCGAAG-GGGCAAAUACAUAGGUGGAUCCCCCGUGCCA-GGCAAACCAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAUGGGGGGU-- .(....)((((((((((....-))(......).))))))))((((((((..((-((((((((...(((((((...))))))).(((....)))..)))))))))).)))))))).-- ( -48.10, z-score = -2.80, R) >droSec1.super_0 3705668 113 - 21120651 UGGCCACUCCGCUUACCGAAG-GGGCAAAUACAUAGGUGGAUCCCCCGUGCCA-GGCAAACCAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAUGGGGGGU-- .......((((((((((....-))(......).))))))))((((((((..((-((((((((...(((((((...))))))).(((....)))..)))))))))).)))))))).-- ( -47.60, z-score = -2.33, R) >droAna3.scaffold_13337 5792771 102 - 23293914 ------CUCUCGUUACCGAGGUGGGCA---AAAUAAAUUGAUUCGCCGGCUUACUACGGCCCAACUACAAUG----GCCAAGAGCGUUGUCAAAUGGGCUCGGCGGUUGGGUGGU-- ------..((((....))))((.(((.---..............))).)).....((.((((((((......----(((..((((.(........).))))))))))))))).))-- ( -32.06, z-score = -0.16, R) >consensus UGGCAACUCCGCUUACCGAAG_GGGCAAAUAAAUAGAUGGAUCCCCCGUGCCA_GGCAAACCAAGUACAAUGCUUCGUUGUGGUUGUUGUCAAAGGGUUUGCCUGAAUGGGGGGU__ .......((((((((((......))........))))))))((((((((..((.((((((((....((((((((.......)).)))))).....)))))))))).))))))))... (-27.08 = -27.70 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:48 2011