| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,469,350 – 11,469,449 |
| Length | 99 |
| Max. P | 0.500000 |

| Location | 11,469,350 – 11,469,449 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.63 |
| Shannon entropy | 0.42645 |
| G+C content | 0.35750 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -8.76 |
| Energy contribution | -9.47 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
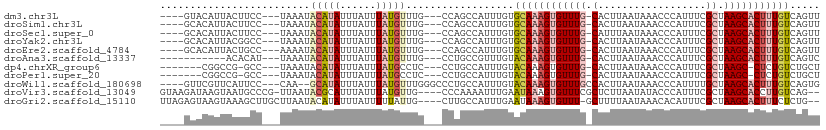
>dm3.chr3L 11469350 99 - 24543557 ----GUACAUUACUUCC---UAAAUACAUAUUUAUUUAUGUUUG---CCAGCCAUUUGUGCAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU ----(((((........---.....(((((......)))))...---.........)))))((((((((((-(..................)).)))))))))....... ( -16.68, z-score = -1.28, R) >droSim1.chr3L 10862817 99 - 22553184 ----GCACAUUACUUCC---UAAAUACAUAUUUAUUUAUGUUUG---CCAGCCAUUUGUGCAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU ----(((((........---.....(((((......)))))...---.........)))))((((((((((-(..................)).)))))))))....... ( -18.68, z-score = -1.82, R) >droSec1.super_0 3677541 99 - 21120651 ----GCACAUUACUUCC---UAAAUACAUAUUUAUUUAUGUUUG---CCAGCCAUUUGUGCAAAGUGUUUG-CAUUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU ----(((((........---.....(((((......)))))...---.........)))))((((((((((-(..................)).)))))))))....... ( -18.68, z-score = -1.88, R) >droYak2.chr3L 11508834 99 - 24197627 ----GCACAUUACGGCC---UAAAUACAUAUUUAUUUAUGUUUG---CCAGCCAUUUGUGCAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU ----(((((....(((.---.....(((((......)))))...---...)))...)))))((((((((((-(..................)).)))))))))....... ( -23.97, z-score = -2.65, R) >droEre2.scaffold_4784 11479266 99 - 25762168 ----GCACAUUACUGCC---AAAAUACAUAUUUAUUUAUGUUUG---CCAGCCAUUUGUGCAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU ----(((((...(((.(---((...(((((......))))))))---.))).....)))))((((((((((-(..................)).)))))))))....... ( -21.97, z-score = -2.02, R) >droAna3.scaffold_13337 5755304 92 - 23293914 -----------ACACAU---UAAAUACAUAUUUAUUUAUGUUUG---CCUGCCGUUUGUACAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUC -----------......---.....(((((......)))))...---........(((.((((((((((((-(..................)).)))))))))))))).. ( -16.37, z-score = -1.53, R) >dp4.chrXR_group6 3119190 94 + 13314419 -------CGGCCG-GCC---UAAAUACAUAUUUAUUUAUGCCUC---CCUGCCAUUUGUACAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGC-CUCUGUCUGCU -------.(((.(-((.---((((((......)))))).)))..---...............(((((....-)))))...................))-).......... ( -16.40, z-score = -0.80, R) >droPer1.super_20 1743084 94 + 1872136 -------CGGCCG-GCC---UAAAUACAUAUUUAUUUAUGCCUC---CCUGCCAUUUGUACAAAGUGUUUG-CACUUAAUAAACCCAUUUCGCUAAGC-CUCUGUCUGCU -------.(((.(-((.---((((((......)))))).)))..---...............(((((....-)))))...................))-).......... ( -16.40, z-score = -0.80, R) >droWil1.scaffold_180698 1968883 101 - 11422946 ----GUUCGUUCAUUCC---CAA--GCAUAUUUAUUUAUGUUUGGGCCCUGCCAUUUGUACAAAGUGUUUGCCACUUAAUAAACCCAUUUUGCUAAGCACUUUGUCAGUG ----....((.....((---(((--(((((......))))))))))....))...(((.((((((((((((.((...(((......))).)).))))))))))))))).. ( -27.20, z-score = -4.04, R) >droVir3.scaffold_13049 1917027 103 + 25233164 GUAAGAUAAGUAAUGCCCG-UUAAUACGCAUUUAUUUAUGUUG----CCCAAAAUUUGAAUAAAGUGUUUCGCUCUUAAUAUACCCAUUUCGCUAAGCACCUUGUCAG-- ((((.(((((((((((..(-(....))))).)))))))).)))----)........(((.(((.((((((.((..................)).)))))).)))))).-- ( -18.77, z-score = -2.33, R) >droGri2.scaffold_15110 14309319 103 - 24565398 UUAGAGUAAGUAAAGCUUGCUUAAUACAUAUUUAUUUUUAUUG----CUUGCCAUUUGAAUAAAGUGUUU-GCUUUUAAUAAACACAUUUCGCUAAGCACUUUCUCUG-- .(((((.((((...((((((..........(((((((..((.(----....).))..)))))))((((((-(.......))))))).....)).)))))))).)))))-- ( -20.80, z-score = -1.34, R) >consensus ____GCACAUUACUGCC___UAAAUACAUAUUUAUUUAUGUUUG___CCAGCCAUUUGUACAAAGUGUUUG_CACUUAAUAAACCCAUUUCGCUAAGCACUUUGUCAGUU .........................(((((......)))))..................((((((((((((.(..................).))))))))))))..... ( -8.76 = -9.47 + 0.71)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:41 2011