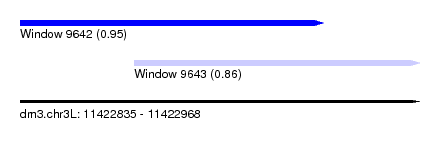
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,422,835 – 11,422,968 |
| Length | 133 |
| Max. P | 0.950147 |

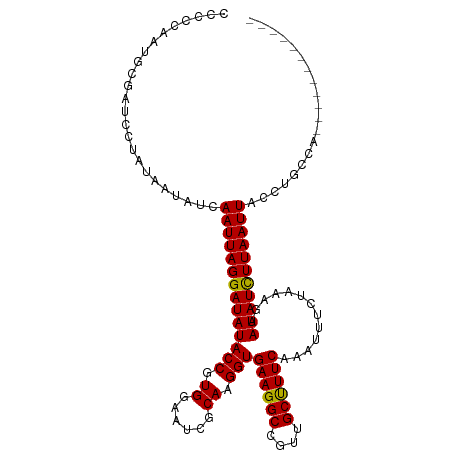
| Location | 11,422,835 – 11,422,936 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.14 |
| Shannon entropy | 0.27501 |
| G+C content | 0.37934 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.26 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11422835 101 + 24543557 CCCCGAAUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAACACAAGGUGAAGGCCAUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUACCUGCCA------------ ........(((((........)))(((((((((((...(((....)))...((((((((....))))))))...........)))))))))))....))..------------ ( -27.10, z-score = -3.66, R) >droEre2.scaffold_4784 11429390 98 + 25762168 CCCA---UGAGAUCCCAUAAUAUCAAUUAGGAUAUACCGUGGAAUAACAAGGUGAAGGCCGUUGCUUUCAAAUUCUUAAGGCAUAUUUUAAUUACCUGCUG------------ ....---.................(((((((((((.((..(((((.......(((((((....))))))).)))))...)).)))))))))))........------------ ( -19.20, z-score = -0.42, R) >droYak2.chr3L 11462029 113 + 24197627 CCCAAAAUGAGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAAAGCAAGGUGAAGGCCGUUGCCUUCAAAAUCUUAAAGCAUAUUUUAAUUACCUGCCAAUAUCUCAUAUA ......(((((((((....(((((......))))).....)))...(((.(((((((((....))))))......((((((....))))))..)))))).....))))))... ( -26.10, z-score = -2.61, R) >droSec1.super_0 3631830 99 + 21120651 --CCCAUUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUAGCUGCCA------------ --......(((((........)))(((((((((((.(..(((((.(....)((((((((....))))))))..)))))..).))))))))))).)).....------------ ( -27.00, z-score = -2.68, R) >droSim1.chr3L 10817111 99 + 22553184 --CCCAAUGCGAUCCUACAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUAGCUGCCA------------ --.....((((((.((((.(((((......)))))...)))).)))))).(((((((((....)))))).........(((.....)))........))).------------ ( -24.50, z-score = -2.01, R) >consensus CCCCCAAUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUACCUGCCA____________ ........................((((((((((((((.((......)).)))((((((....)))))).............))))))))))).................... (-18.66 = -18.26 + -0.40)

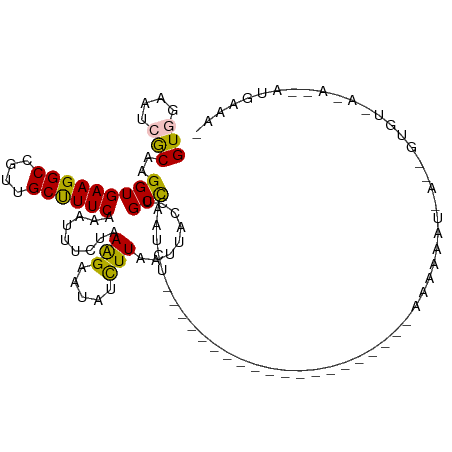
| Location | 11,422,873 – 11,422,968 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.19 |
| Shannon entropy | 0.53829 |
| G+C content | 0.35325 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -9.96 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 11422873 95 + 24543557 GUGGAAACACAAGGUGAAGGCCAUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUGC--------------------AAGAAAUUAGUGUGUGAAAGCAUGAAAC (((....)))..(((....)))..(((((((((((..((((((.....))).....(.(((....))--------------------).)....))).)))))))))))...... ( -26.60, z-score = -3.19, R) >droEre2.scaffold_4784 11429425 95 + 25762168 GUGGAAUAACAAGGUGAAGGCCGUUGCUUUCAAAUUCUUAAGGCAUAUUUUAAUUACCUGCUGAU--------------------UAUAUAAAUGACAGUGUAAAAAAAUGAAAU ......(((.((..(((((((....)))))))..)).))).....((((((..((((..((((.(--------------------(((....)))))))))))).)))))).... ( -14.40, z-score = 0.11, R) >droYak2.chr3L 11462067 115 + 24197627 GUGGAAAAGCAAGGUGAAGGCCGUUGCCUUCAAAAUCUUAAAGCAUAUUUUAAUUACCUGCCAAUAUCUCAUAUAACUACUGUAUUAUACAAAUGAGAGUGUGAGAACAUGAAAU (((.....(((.(((((((((....))))))......((((((....))))))..)))))).....((((((((..((..((((...))))....)).)))))))).)))..... ( -23.70, z-score = -1.42, R) >droSec1.super_0 3631866 73 + 21120651 GUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUCU--------------------AUAAAC---------------------- (((((...(((..((((((((....)))))))).......(((.....))).......)))...)))--------------------))....---------------------- ( -16.30, z-score = -1.68, R) >droSim1.chr3L 10817147 73 + 22553184 GUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUCU--------------------AUAAAC---------------------- ((((((((....)))...(((.((((.((......(((....)))......)).)))).)))..)))--------------------))....---------------------- ( -14.30, z-score = -1.05, R) >consensus GUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUCU____________________AAAAAAU_A__GUGU_A_A__AUGAAA_ (((....)))..(((((((((....)))))).........(((.....)))........)))..................................................... ( -9.96 = -9.40 + -0.56)

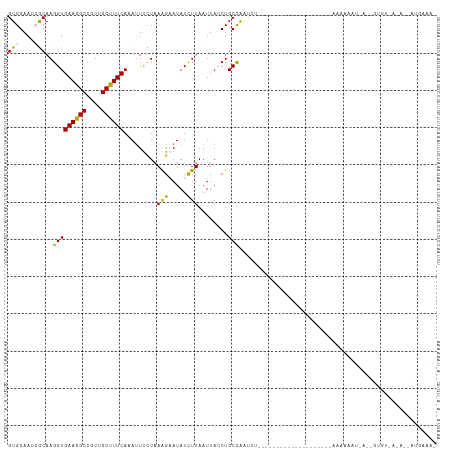
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:38 2011