| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,421,581 – 11,421,764 |
| Length | 183 |
| Max. P | 0.981687 |

| Location | 11,421,581 – 11,421,764 |
|---|---|
| Length | 183 |
| Sequences | 4 |
| Columns | 184 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.27378 |
| G+C content | 0.35859 |
| Mean single sequence MFE | -46.36 |
| Consensus MFE | -34.75 |
| Energy contribution | -34.94 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
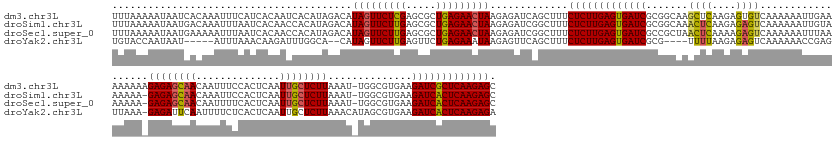
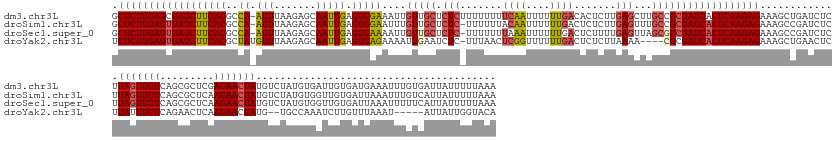
>dm3.chr3L 11421581 183 + 24543557 UUUAAAAAUAAUCACAAAUUUCAUCACAAUCACAUAGACAUAGUUCUCGAGCGCUGAGAACUAAGAGAUCAGCUUUCUCUUGAGUGAUCGCGGCAAGCUCAAGAGUGUCAAAAAAUUGAAAAAAAAGAGAGCAACAAUUUCCACUCAAUUGCUCUUAAAU-UGGCGUGAAGAUCGCUCAAGAGC .................(((((.......((.....))..(((((((((.....))))))))).))))).......((((((((((((((((.((((((....))).((((....)))).......((((((((..............))))))))...)-)).)))...))))))))))))). ( -47.14, z-score = -2.11, R) >droSim1.chr3L 10815828 182 + 22553184 UUUAAAAAUAAUGACAAAUUUAAUCACAACCACAUAGACAUAGUUCUUGAGCGCUGAGAACUAAGAGAUCGGCUUUCUCUUGAGUGAUCGCGGCAAACUCAAGAGAGUCAAAAAAUUGUAAAAAA-GAGAGCAACAAAUUCCACUCAAUUGCUCUUAAAU-UGGCGUGAAGAUCACUCAAGAGC ........................................((((((((.......)))))))).............((((((((((((((((.(((((((....)))).................-((((((((..............))))))))...)-)).)))...))))))))))))). ( -45.34, z-score = -2.38, R) >droSec1.super_0 3630556 182 + 21120651 UUUAAAAAUAAUGAAAAAUUUAAUCACAACCACAUAGACAUAGUUCUUGAGCGCUGAGAACUAAGAGAUCGGCUUUCUCUUGAGUGAUCGCCGCUAACUCAAAAGAGUCAAAAAAUUUAAAAAAA-GAGAGCAACAAUUUUCACUCAAUUGCUCUUAAAU-UGGCGUGAAGAUCACUCAAGAGC ........................................((((((((.......)))))))).............(((((((((((((..(((((((((....)))).................-((((((((..............))))))))....-)))))....))))))))))))). ( -45.94, z-score = -3.11, R) >droYak2.chr3L 11460716 172 + 24197627 UGUACCAAUAAU-----AUUUAAACAAGAUUUGGCA--CAUAGUUCUUGAGUUCUGAGAAAUAAGAGUUCAGCUUUCUCUUGAGUGAUCGCG----UUUUAAGAGAGUCAAAAAACCGAGUUAAA-GAGAUUCAAUUUUCUCACUCAAUUGCUCUUAAACAUAGCGUGAAGAUCACUCAAGAGA (((.((((....-----.............)))).)--))......((((..(((........)))..))))...(((((((((((((((((----(((((((((.((......)).((((....-((((........)))))))).....))))))))....))))...)))))))))))))) ( -47.03, z-score = -2.85, R) >consensus UUUAAAAAUAAUGACAAAUUUAAUCACAACCACAUAGACAUAGUUCUUGAGCGCUGAGAACUAAGAGAUCAGCUUUCUCUUGAGUGAUCGCGGCAAACUCAAGAGAGUCAAAAAAUUGAAAAAAA_GAGAGCAACAAUUUCCACUCAAUUGCUCUUAAAU_UGGCGUGAAGAUCACUCAAGAGC ........................................(((((((((.....))))))))).............(((((((((((((.......((((....))))..................((((((((..............))))))))..............))))))))))))). (-34.75 = -34.94 + 0.19)
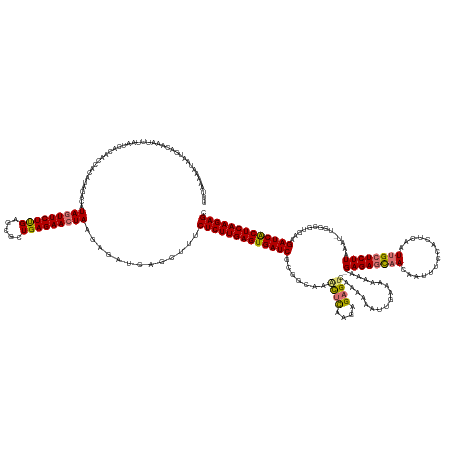
| Location | 11,421,581 – 11,421,764 |
|---|---|
| Length | 183 |
| Sequences | 4 |
| Columns | 184 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.27378 |
| G+C content | 0.35859 |
| Mean single sequence MFE | -49.22 |
| Consensus MFE | -31.83 |
| Energy contribution | -33.45 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
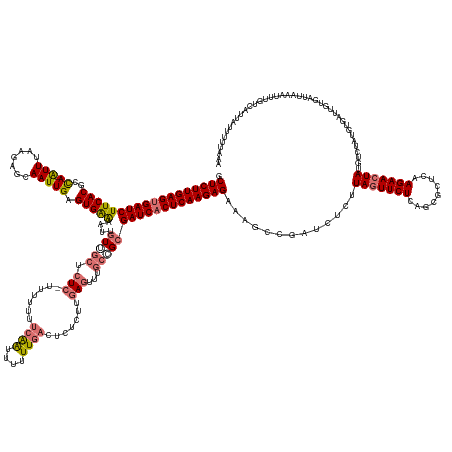
>dm3.chr3L 11421581 183 - 24543557 GCUCUUGAGCGAUCUUCACGCCA-AUUUAAGAGCAAUUGAGUGGAAAUUGUUGCUCUCUUUUUUUUCAAUUUUUUGACACUCUUGAGCUUGCCGCGAUCACUCAAGAGAAAGCUGAUCUCUUAGUUCUCAGCGCUCGAGAACUAUGUCUAUGUGAUUGUGAUGAAAUUUGUGAUUAUUUUUAAA ((((..(((.(............-.....((((((((..(((....)))))))))))........((((....))))).)))..))))....(((((((((..((((((.......))))))(((((((.......)))))))........)))))))))........................ ( -47.60, z-score = -0.96, R) >droSim1.chr3L 10815828 182 - 22553184 GCUCUUGAGUGAUCUUCACGCCA-AUUUAAGAGCAAUUGAGUGGAAUUUGUUGCUCUC-UUUUUUACAAUUUUUUGACUCUCUUGAGUUUGCCGCGAUCACUCAAGAGAAAGCCGAUCUCUUAGUUCUCAGCGCUCAAGAACUAUGUCUAUGUGGUUGUGAUUAAAUUUGUCAUUAUUUUUAAA .(((((((((((((.....((.(-(((((((((.....(((..(......)..)))..-.......(((....)))...)))))))))).))...)))))))))))))..(((((......(((((((.........)))))))........)))))(((((.......))))).......... ( -49.44, z-score = -2.37, R) >droSec1.super_0 3630556 182 - 21120651 GCUCUUGAGUGAUCUUCACGCCA-AUUUAAGAGCAAUUGAGUGAAAAUUGUUGCUCUC-UUUUUUUAAAUUUUUUGACUCUUUUGAGUUAGCGGCGAUCACUCAAGAGAAAGCCGAUCUCUUAGUUCUCAGCGCUCAAGAACUAUGUCUAUGUGGUUGUGAUUAAAUUUUUCAUUAUUUUUAAA .(((((((((((((.....(((.-.....((((((((..(((....))))))))))).-..............(((((((....))))))).))))))))))))))))..(((((......(((((((.........)))))))........)))))((((.........)))).......... ( -51.74, z-score = -3.12, R) >droYak2.chr3L 11460716 172 - 24197627 UCUCUUGAGUGAUCUUCACGCUAUGUUUAAGAGCAAUUGAGUGAGAAAAUUGAAUCUC-UUUAACUCGGUUUUUUGACUCUCUUAAAA----CGCGAUCACUCAAGAGAAAGCUGAACUCUUAUUUCUCAGAACUCAAGAACUAUG--UGCCAAAUCUUGUUUAAAU-----AUUAUUGGUACA ((((((((((((((.....((....(((((((......((((.((((((((((.....-......)))))))))).))))))))))).----.))))))))))))))))...((((...........)))).............((--((((((....(((....))-----)...)))))))) ( -48.10, z-score = -3.62, R) >consensus GCUCUUGAGUGAUCUUCACGCCA_AUUUAAGAGCAAUUGAGUGAAAAUUGUUGCUCUC_UUUUUUUCAAUUUUUUGACUCUCUUGAGUUUGCCGCGAUCACUCAAGAGAAAGCCGAUCUCUUAGUUCUCAGCGCUCAAGAACUAUGUCUAUGUGAUUGUGAUUAAAUUUGUCAUUAUUUUUAAA .(((((((((((((....((.((.(((((((((.....((((((......)))))).........((((....))))..))))))))).)).)).))))))))))))).............(((((((.........)))))))........................................ (-31.83 = -33.45 + 1.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:37 2011