| Sequence ID | dm3.chr3L |
|---|---|
| Location | 11,420,574 – 11,420,673 |
| Length | 99 |
| Max. P | 0.513828 |

| Location | 11,420,574 – 11,420,673 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.51564 |
| G+C content | 0.42977 |
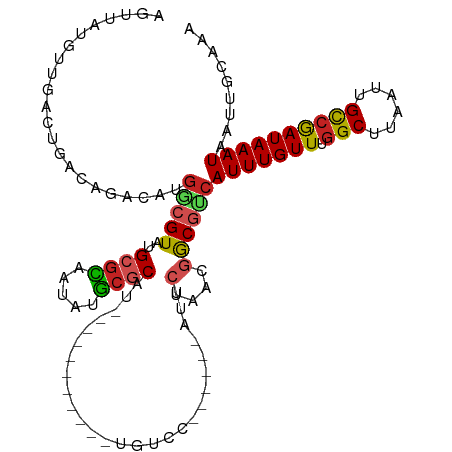
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.35 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
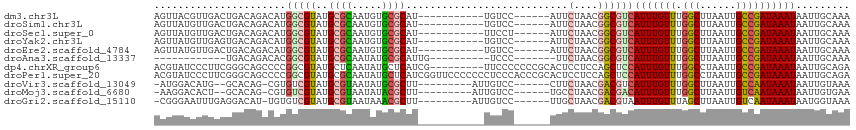
>dm3.chr3L 11420574 99 - 24543557 AGUUACGUUGACUGACAGACAUGGCGUAUGCGCAAUGUGCGCAU-----------UGUCC------AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA .(((..(((....)))(((.((((((.((((((.....))))))-----------)).))------)))))))).(((..(((((((.(((......))))))))))...)))... ( -28.10, z-score = -0.81, R) >droSim1.chr3L 10814851 99 - 22553184 AGUUAUGUUGACUGACAGACAUGGCGUAUGCGCAAUGUGCGCAU-----------UGUCC------AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA .((((((((....))))((.((((((.((((((.....))))))-----------)).))------)))))))).(((..(((((((.(((......))))))))))...)))... ( -28.70, z-score = -1.08, R) >droSec1.super_0 3629626 99 - 21120651 AGUUAUGUUGACUGACAGACAUGGCGUAUGCGCAAUGUGCGCAU-----------UUCCU------AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA .((((((((........))))))))..((((((.....))))))-----------.....------.........(((..(((((((.(((......))))))))))...)))... ( -27.20, z-score = -1.11, R) >droYak2.chr3L 11459557 99 - 24197627 AGUUAUGUUGAGUGACAGACAUGGCGUAUGCGCAAUGUGCGCAU-----------UGUCC------AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA .(((((.....)))))(((.((((((.((((((.....))))))-----------)).))------)))))....(((..(((((((.(((......))))))))))...)))... ( -29.70, z-score = -1.23, R) >droEre2.scaffold_4784 11426799 99 - 25762168 AGUUAUGUUGACUGACAGACAUGGCGUAUGCGCAAUGUGCGCAU-----------UGUCC------AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA .((((((((....))))((.((((((.((((((.....))))))-----------)).))------)))))))).(((..(((((((.(((......))))))))))...)))... ( -28.70, z-score = -1.08, R) >droAna3.scaffold_13337 5702872 87 - 23293914 ------------UGACAGACACGGCGUAUGCGCAAUAUGCGCAUUG----------UCCC-------UUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA ------------....(((...((((.((((((.....))))))))----------))..-------.)))....(((..(((((((.(((......))))))))))...)))... ( -24.80, z-score = -1.27, R) >dp4.chrXR_group6 3032543 107 + 13314419 ACGUAUCCCUUCGGGCAGCCCCGGCGUAUGCUCAAUAUGCUCAUCG---------UUCCCCCCCGCACUCCUCCAGCUCCAUUUGUUUGGCCUAAUUGCCGAUAAAUAAUUGCAGA ............(((.(((...(((((((.....)))))))....)---------)).)))..............((...(((((((.(((......))))))))))....))... ( -21.70, z-score = 0.21, R) >droPer1.super_20 1643282 116 + 1872136 ACGUAUCCCUUCGGGCAGCCCCGGCGUAUGCGCAAUAUGCUCAUCGGUUCCCCCCCUCCCACCCGCACUCCUCCAGCUCCAUUUGUUUGGCCUAAUUGCCGAUAAAUAAUUGCAGA ............(((.((((..(((((((.....)))))))....)))).)))......................((...(((((((.(((......))))))))))....))... ( -24.70, z-score = 0.35, R) >droVir3.scaffold_13049 1737370 97 + 25233164 -AUGGACAUG--GCACAG-CGUGUCGUAUGCGUAAUAUGCGCUU---------AUUGUCC------CUUCUAACGACGUCAUUUGUUUGGCUUAAUUGCCAAUAAAUAAUUGUAAA -..(((((((--((((..-.)))))))..((((.....))))..---------...))))------.........(((..(((((((.(((......))))))))))...)))... ( -25.10, z-score = -1.36, R) >droMoj3.scaffold_6680 6658047 97 + 24764193 -AAGGACACU--GCACAG-CGUGUCGUAUGCGUAAUAUACGCUU---------AUUGUCC------UGCCUAACGACGACAUUUGUUUGGCUUAAUUGUCAAUAAAUAAUUGUGAA -.((((((.(--(...((-(((((((....))....))))))))---------).)))))------)(((.(((((......))))).))).(((((((......))))))).... ( -23.90, z-score = -1.08, R) >droGri2.scaffold_15110 14153290 99 - 24565398 -CGGGAAUUUGAGGACAU-UGUGUCGUAUGCGUAAUAAACGCUU---------AUUGUCC------UUGCUAACGACGUAAUUUGUUUAGCUUAAUUGUCAAUAAAUAAUGGUAAA -.((.((((.(((((((.-......(((.((((.....)))).)---------)))))))------))((((((((......))).)))))..)))).))................ ( -19.51, z-score = -0.38, R) >consensus AGUUAUGUUGACUGACAGACAUGGCGUAUGCGCAAUAUGCGCAU___________UGUCC______AUUCUAACGGCGUCAUUUGUUUGGCUUAAUUGCCGAUAAAUAAUUGCAAA ......................(((((..((((.....))))...........................(....))))))(((((((.(((......))))))))))......... (-15.47 = -15.35 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:18:34 2011